This is the second attempt at discussing a very interesting paper which has been hurt by an editor error in publication (a key informative element, table 2, has its columns all swapped).
I realized that something looked quite wrong and notified the authors, who are now awaiting for PLoS to correct the problem. In the meantime they have been so kind as to provide me with a copy of the original PDF manuscript so I could properly collate the haplogroup data and share it with readers of this blog.
Ganesh Prasad Arun Kumar et al., Population Differentiation of Southern Indian Male Lineages Correlates with Agricultural Expansions Predating the Caste System. PLoS ONE 2012. Open access → LINK [doi:10.1371/journal.pone.0050269]
As I said back in the day:
The authors took special interest into sampling tribes, some of which
are still foragers and a reference for all kind of anthropological
research of South Asia, all Eurasia and even beyond. They also sorted
the various populations into groups or classes based on socio-economic
reality (and language in some cases) rather than the, arguably
overrated, varna (caste) system.
The categories used are:
- HTF - Hill Tribe Forager (foragers of Tamil or Malayalam language)
- HTK - Hill Tribe Kannada (foragers of Kannada language)
- HTC - Hill Tribe Cremation (tribals who cremate their dead, not sure if silviculturalists)
- SC - Scheduled Castes (castes traditionally discriminated against, Dalits)
- DLF - Dry Land Farmers
- AW - Artisan and Warrior related castes
- BRH - Brahmin-related castes with irrigation farming economy
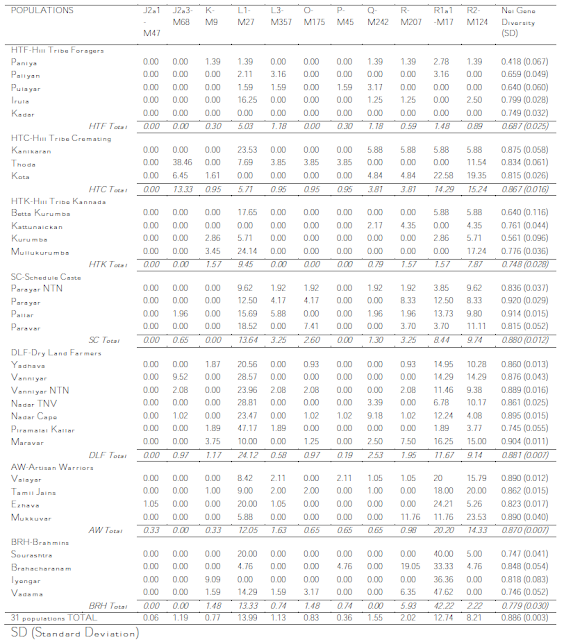
And, as I said then, the bulk of the data is in table 2, which I have the privilege of sharing with you as it really is (in two blocs, as it was in the PDF):
And now finally I can get to discuss the details with the certainty of talking about real data.
Haplogroup C
As the authors note, 90% (66/74) of all the C-M130 samples belong to C5 (M356), while the rest (8/74) tested negative for both C5 and C3 (M217), so I guess we are here before at leas one other subhaplogroup of C (because the likelihood of being Japanese C1 or Australasian C2 or C4 is practically zero).
The eight C* individuals are scattered (table S1) among several groups (all of which also display C5, as well as F*) but notably concentrated among the Piramalai Kallar (4/5 within C), which are a DLF group (corrected upon comment).
Besides C*, which may well be a remnant of either the early Eurasian expansion or of the first backflows from SE Asia (a likely not-so-likely candidate for the origin of macro-haplogroup C), the very notable presence of C5 among tribals and some farmers may well indicate that the origin of C5 is in South Asia, even if the clade also has some presence in Central and West Asia.
Haplogroup C has a high variance in this study (0.80), greatest among DLF (0.89) and HTF (0.81).
(Update: see also appendix below).
(Update: see also appendix below).
Haplogroup E
As we should expect, this lineage of African origin (with important presence in West Eurasia) is only found at low levels among farmers (DLF). It may well be a remnant of early Neolithic flows, being strongly linked with Neolithic in the case of Europe for example.
Paragroup F*
The most striking thing about Paragroup F*, i.e. F(xG,H,J,K), is that it is found at such high numbers and very especially so among the hunter-gatherers, where it is often the main lineage (or lineages). It is also important among dry land farmers and the Valayar (AW class) but it is rare to non-existent among the other caste groups, which may represent relatively recent arrivals.
Something that this confirms, along with other older data about F basal diversity, is that the main Eurasian Y-DNA haplogroup, which is of course F itself, coalesced necessarily in South Asia.
Said that, I cannot underline enough how relevant is to find rare F sublineages (i.e. F* - so rare that have not even been properly identified by downstream markers yet) among the last forager peoples of South Asia, often as dominant clade.
Haplotype neighbor-joining exercise was performed however, indicating founder effects (possible new haplogroups to be yet described) among tribals:
However it is also obvious that there is a lot of diversity as well. In fact, paragroup F* does have a high variance in Tamil Nadu (0.81), being highest again among the DLF class (0.85).
Haplogroup G
Haplogroup G does exist in South Asia and this paper makes it evident. More so, its distribution in Tamil Nadu includes some foragers and other tribals, although it is more common among "Neolithic" classes.
Among these the Ivayengar (BRH) show almost 27% (3/11), however other BRH populations do not show any G, while the DLF ones instead all have relevant G. Therefore this lineage may tentatively be associated in Tamil Nadu with the Neolithic.
Haplogroup G, suggested by the authors to be a Neolithic arrival, has an strikingly high variance in Tamil Nadu (0.83) with top level among the following classes: AW (1.05), SC (0.94) and BRH (0.82).
Even if the distribution corresponds well with a Neolithic inflow the diversity is surprisingly high and it tells me that more research is needed about this lineage in South Asia. After all it is one of the basal descendants of F, whose coalescence took place no doubt in the subcontinent.
Haplogroup H
Haplgroup H is of course very common in Tamil Nadu but it must be noticed that it is concentrated in the H1(xH1a) category, as well as some notable H(xH1,H2), which tends to weight in favor of a southern ultimate origin of this important South Asian clade (as also proposed in the recent study on the Roma People).
H* is distributed among many populations, the only class fully excluded being the BHR one, which is generally considered to be a recent historical arrival from the North (mostly confirmed by genetics). Some tribes have the highest values but then some others totally lack it.
H* has extremely high variance levels in Tamil Nadu (1.33), being highest among the SC class (1.46), followed by the AW one (1.18) and the DLF one (0.91). This is totally consistent with a South Asian origin of H overall.
H1* is standard issue in all populations. The highest values are among the Kannada-speaking tribals (HTK), followed by cremation-practicing tribals (HTC).
H1a instead is only found in one population at very low levels, strongly suggesting that this clade is not from the region. H2 instead is found at low levels among many groups.
Unlike H*, H1 and H2 have rather low diversity levels in Tamil Nadu: 0.41 and 0.59 respectively.
Haplogroup J
J(xJ2) is found at anecdotal levels in a couple of lower class populations (one tribal and the other SC). It would be particularly interesting if we knew it is not J1 as well - but we don't.
Most is J2(xJ2a) although J2a3 is also important among several populations.
It is generally believed that J in South Asia is of Neolithic origin and I will not question it but still... notice how important it is among several tribal foragers: >4% in four tribes, levels on average similar to those of farmers and Brahmins.
J2* is rather high in diversity (0.73), notably among the AW class (1.0), while J2a3 is very low instead (0.29).
Paragroup K(xL,R)
Or if you wish paragroups K* and P*, as well as haplogroups O and Q.
The always interesting K* is found at low levels among some tribals and most DLF populations. However the peak is among Viyengar Brahmins. May it be haplogroup T?, L2?
O in this area is almost for sure O2a brought by Austroasiatic-speaking rice farming tribes in the Neolithic. It is found at low levels in some groups, including the Thoda "cremation tribals" (who look quite "Neolithic" also because of their high levels of J2).
P(xQ,R), which is most common towards Bengal, is found in Tamil Nadu at low levels among diverse populations. On first impression I'd say it's also a Neolithic influence although, of course, in the wider subcontinental region it must be much much older.
Q is found at low levels in diverse populations being maybe somewhat more common among the Scheduled Castes class.
Haplogroup L
Haplogroup L is an important South Asian lineage with penetration in West and Central Asia and a center of gravity around Sindh (Pakistan), although it is also very common in West and South India.
In Tamil Nadu L1a (L1 in the table) is common among nearly all sampled populations with peak among the dry land farmers' class.
Instead L1c (formerly L3) is relatively rare, peaking among the Scheduled Castes class. No mention is made of any other L.
Both clades show low variance in the region (0.41 and 0.22 respectively), consistent with their origin being further North.
Haplogroup R
R(xR1a1,R2) is found in several populations at non-negligible levels: near 5% among some tribals, 8% among the Parayar (SC) and the Maravar (DLF), also 12% Mukkuvar (AW) and as much as 19% among some Brahmins (the Brahacharanam who are also high in P*). This could well be R*, R1*, R1a*, R1b, etc. and indicates in my understanding target populations for future research on the hot topic of the ultimate origins of R1 and R1a (see also here).
R* shows clearly high variance: 0.97 on average, being highest among the DLF class (1.25), followed by the BRH class (0.99)
R1a1, which may well be related to Indoeuropean expansion (or just Neolithic or whatever, better resolution is needed especially in Asia) is found at very high levels among the BRH class (45%), followed by the AW one (20%) other classes show near 10% except the hunter-gatherers (HTF and HTK) who have only anecdotal presence of this lineage.
R1a1 shows rather low variance (0.41), rather confirming its immigrant origin from North India (incl. maybe Pakistan, Bangla Desh, Nepal...). All classes are similar for this value.
R2 (a South Asian lineage with occasional offshoots into West and Central Asia) is common in all groups except the HTF class. The highest levels (c. 15% avg.) are among cremating tribals and artisan/warrior classes. I'd say that with the likely origin of R2 somewhat to the North of this region, it seems normal that Kannada-speaking tribals (HTK, who must be immigrants from Karnataka or at least strongly influenced by this other Dravidian country's culture) have lots of it, while the more locally native HTF almost lack it instead.
R2 shows mid-level diversity on average (0.65) but the HTK class displays very high diversity for this lineage (1.05).
Different interpretations
Notice that my take and that of the authors on the autochthonous nature of each of the lineages may vary or be debatable. They say the following:
The geographical origins of many of these HGs are still debated. However, the associated high frequencies and haplotype variances of HGs H-M69, F*-M89, R1a1-M17, L1-M27, R2-M124 and C5-M356 within India, have been interpreted as evidence of an autochthonous origins of these lineages during late Pleistocene (10–30 Kya), while the lower frequency within the subcontinent of J2-M172, E-M96, G-M201 and L3-M357 are viewed as reflecting probable gene flow introduced from West Eurasian Holocene migrations in the last 10 Kya [6], [7], [16], [23]. Assuming these geographical origins of the HGs to be the most likely ones, the putatively autochthonous lineages accounted for 81.4±0.95% of the total genetic composition of TN populations in the present study.
Mostly our differences stem on my doubts about the real origins of R1a1 (which could well be West Asian by origin) and that I imagined L1c (aka L3) as native from South Asia (uncertain now admittedly). But otherwise I agree. The hottest issue is no doubt the origin of R1a or R1a1, still unsolved.
PC Analysis
A quick visual understanding of the relations between the different classes can be obtained from figure 2:
Check table 1 for population codes but essentially: squares are tribes and circles caste populations; red are the HTF class, green the HTK and yellow the Brahmin-related groups (BRH).
These are the outliers: all the rest, including HTC, cluster together near the (0,0) coordinates.
It is also clearly indicated in Fig. 2A how R1a1, H1 and F* are the strongest defining markers.
Old structure
As always, take age estimates, also provided, with utmost caution and distrust. However I must mention that the main conclusion of the authors is that the haplogroup structure in the region pre-dates the introduction of the caste system as such and is, in their opinion, of Neolithic age.
Much of the discussion below has been on the origins of haplogroup C. I have been pointed to Hammer 2006 and this haplotype NJ tree (fig 4d) of what was known back in the day as C* and C1. At that time neither Australian C4 nor Asian C5 had been described yet. However Wallacean/Melanesian/Polynesian C2 and NE Asian and Native American C3 are not shown here.
Annotations (C1, C4 and root?) by me:
__________________________ . __________________________
Appendix (update Dec 2):
Much of the discussion below has been on the origins of haplogroup C. I have been pointed to Hammer 2006 and this haplotype NJ tree (fig 4d) of what was known back in the day as C* and C1. At that time neither Australian C4 nor Asian C5 had been described yet. However Wallacean/Melanesian/Polynesian C2 and NE Asian and Native American C3 are not shown here.
Annotations (C1, C4 and root?) by me:
Maybe even more interesting is Fig. 3 from Redd 2002, which shows the whole C haplogroup tree and clearly annotates the likely root (branch to haplogroup B):
While C4 is not obvious here, the fact that South Asian (Indian subcontinent) C* is central to all the haplogroup is again underlined.
The protuberance to the top might be C5, while the one to the bottom may well correspond with the SE Asian cluster above, at least partly. The differences underline the limitations of this STR-based method alone to infer real phylogenies - but it is anyhow much better than nothing.



A couple of observations:
ReplyDelete" Something that this confirms, along with other older data about F basal diversity, is that the main Eurasian Y-DNA haplogroup, which is of course F itself, coalesced necessarily in South Asia"
Not really necessarily so. It possibly merely indicates that F was able to diversify considerably once it entered the subcontinent, especially when we consider:
"Haplogroup G, suggested by the authors to be a Neolithic arrival ... After all it is one of the basal descendants of F, whose coalescence took place no doubt in the subcontinent".
But G almost certainly did not coalesce 'in the subcontinent'. Therefore your statement here concerning F is the result of what you have already decided what you want to see. Concerning another basal F haplogroup you write:
"H* is distributed among many populations, the only class fully excluded being the BHR one, which is generally considered to be a recent historical arrival from the North (mostly confirmed by genetics)".
So H most probably coalesced somewhere near F's entry point into South Asia. Concerning yet another basal F haplogroup IJ you write:
"It is generally believed that J in South Asia is of Neolithic origin"
Note: no I. So IJ too coalesced outside South Asia, presumably branching off before F had entered the subcontinent.
"P(xQ,R), which is most common towards Bengal"
Which fits the breakup of MNOPS being somewhere at least as far east as Bengal.
"Besides C*, which may well be a remnant of either the early Eurasian expansion or of the first backflows from SE Asia (a likely candidate for the origin of macro-haplogroup C), the very notable presence of C5 among tribals and some farmers may well indicate that the origin of C5 is in South Asia, even if the clade also has some presence in Central and West Asia".
We really need a closer look at C's phylogeny. The high level of C5 in Southern India is very interesting, of course. The low level of C* is interesting as well considering that its presence in the Subcontinent has consistently been used as evidence of the great southern coastal migration. It seems now that unclassified C* is concentrated in SE Asia, which fits 'SE Asia (a likely candidate for the origin of macro-haplogroup C)'.
Off Topic: very interesting abstract - unfortunately behind paywall, so I have not read the paper:
ReplyDeletehttp://www.genetics.org/content/192/3/1065.abstract
I read something about it but mostly a press release on Native Americans blah blah that seems quite unrelated. Razib wrote today that it's not what it seems and certainly nothing new. In fact the paper was published in August and some bloggers already commented on it.
DeleteI agree. The federal money is arguably a sure thirst quencher for the pocket. Besides I like all non polluting people rather than divided families in on the cut.
DeleteWhat do you mean in the last part of the comment? I was about to moderate it but I really don't understand your slang: what is "polluting people" according to you?
DeleteMaju said,
ReplyDelete"The eight C* individuals are scattered (table S1) among several groups (all of which also display C5, as well as F*) but notably concentrated among the Piramalai Kallar (4/5 within C), which should correspond (if I'm correct) to the Kadar (HTF) of the table."
"Piramalai Kallar" is the sixth population from the top in the DLF (Dry Land Farmers) category, following "Nadar Cape" and preceding "Maravar." This sample is reported to have contained 5/53 = 9.43% C-M130 Y-DNA.
"Kadar," on the other hand, is the fifth and final population in the HTF (Hill Tribe Foragers) category, following "Irula." This sample is reported to have contained 3/28 = 10.71% C-M130 Y-DNA.
Long no see.
DeleteAs usual you are right. I will correct that. Thanks.
Maju said,
ReplyDelete"As the authors note, 90% (66/74) of all the C-M130 samples belong to C5 (M356), while the rest (8/74) tested negative for both C5 and C3 (M217), so I guess we are here before at leas one other subhaplogroup of C (because the likelihood of being Japanese C1 or Australasian C2 or C4 is practically zero)."
It has been known ever since the discovery of C5-M356 that the overwhelming majority of C-M130 representatives from the Subcontinent belong to this clade. This finding was published in the same paper that reported the discovery and phylogenetic placement of M356 seven years ago (Sanghamitra Sengupta, Lev A. Zhivotovsky, Roy King et al., "Polarity and Temporality of High-Resolution Y-Chromosome Distributions in India Identify Both Indigenous and Exogenous Expansions and Reveal Minor Genetic Influence of Central Asian Pastoralists"):
"India"
11/728 = 1.51% C5-M356
2/728 = 0.27% C*-M216/RPS4Y(xC1-M8, C2-M38, C3-M217, C4-M210, C5-M356)
11/13 = 85% C5-M356/total C-M216
In other words, the finding that C5-M356 comprises approximately nine tenths of all C-M130/M216 representatives from India is nothing new. However, it is known that representatives of C3-M217 also exist in the northwestern (Pakistan) and northeastern (Nepal) extremities of the Subcontinent, probably reflecting the Mongoloid influence in these peripheral regions.
In regard to the ancestry of C5-M356, it has been known since at least ten years ago (Redd et al. 2002) that, in median-joining networks of Y-STR variation, the majority of C-M130 representatives from the Indian Subcontinent cluster together and are well separated from Oceanic C2-M38, which seems to be the outlier among extant subclades of C-M130. Both Redd et al. and a later similar analysis by Hammer et al. have shown South Asian C5-M356 to be "intermediate" between Japanese C1-M8 and East Asian/American C3-M217 in regard to Y-STR variation, yet each of these clades clusters separately despite being obviously more closely related to one another than any is related to Oceanic C2-M38. C5-M356 is also relatively close to Australian C4-M210, although the latter clade deviates slightly in the direction of the very distinctive C2-M38.
In any case, Japanese C1-M8, East Asian/American C3-M217, Australian C4-M210, and South Asian C5-M356 seem to comprise a major cluster of extant C-M130 Y-chromosomes. As for South Asian C*-M130 Y-DNA, it seems to form a loose cluster with Southeast Asian C*-M130 Y-DNA, and this cluster of Southeast/South Asian C*-M130 seems to have diverged from the C-root around the same time as (and from an ancestor closely related to the ancestor of) Oceanic C2-M38. In other words, the very rare South Asian C*-M130 does not seem to be particularly closely related to the more common South Asian C5-M356; C5-M356 seems to be a recently expanding "successful" ("Neolithic"?) clade relatively closely related to Japanese C1-M8, East Asian/American C3-M217, and Australian C4-M210, whereas South Asian C*-M130 seems to reflect the sparse survivors of a very ancient population that once covered a wide area encompassing Southeast Asia and South Asia (perhaps the "tropical" substrate of the Austroasiatic-speaking peoples?), whose progeny survived and prospered more notably in Austronesian-speaking regions of Oceania.
Considering the fact that C5-M356 Y-DNA also has been found in Central Asia (e.g. Uyghurs) and Southwest Asia, I think it is a bit too early to make any definite claims on its region of origin.
Not sure what your point is but I seriously doubt that C5 could be Neolithic in South Asia (or in general). After all is a basal subclade of C and hence the dispersal should correspond to the period of the Great Asian Expansion (flows and counterflows through Tropical Asia and beyond in the aftermath of the OoA), which should culminate before c. 50 Ka. almost for sure.
DeleteIn a previous discussion (lost in the ocean of comments and counter-comments by now but still in my memory) I proposed that C5 may have been the Y-DNA partner (or an Y-DNA partner together with P) of the backflow of mtDNA N (which looks like coalesced in SE Asia). This was contested by Terry (and someone else?) arguing that all C5 in South Asia was in the North (what is obviously not the case but would argue against coastal back-flow) and pointing out to C5 in Central Asia, as if that would be the route of the lineage.
In understand that this finding of C5 together with C* in the southernmost regions of South Asia, with strong presence among foragers and together with other archaic-looking clades like F* and H* (and unlinked to O, which is the main marker of East Asian Neolithic influence) really brings the matter again to an scenario of very ancient South-SE Asia flows and backflows, what happened necessarily before c. 50 Ka., whatever the "molecular clock" speculations say.
Maju said,
ReplyDelete"In understand that this finding of C5 together with C* in the southernmost regions of South Asia, with strong presence among foragers and together with other archaic-looking clades like F* and H* (and unlinked to O, which is the main marker of East Asian Neolithic influence) really brings the matter again to an scenario of very ancient South-SE Asia flows and backflows, what happened necessarily before c. 50 Ka., whatever the "molecular clock" speculations say."
The problem is that these same populations of foragers also contain representatives of J2-M172, R1a1a-M17, and other clades that are supposed to reflect Neolithic (or post-Neolithic) colonization of the Indian Subcontinent. In addition, as I have explained in my previous comment, C5-M356 seems to be only very distantly related to South/Southeast Asian C*-M130, even more distantly than C5-M356 is related to C1-M8 or C3-M217. I do not see any reason to presume that C5-M356 has diverged from a common ancestor with South/Southeast Asian C*-M130 within the Indian Subcontinent. However, if it turns out that it is actually the case that both C5-M356 and South Asian C*-M130 are autochthonous in India, then it would probably entail that East Asian/American C3-M217, Japanese C1-M8, and even Australian C4-M210 also all originated in India and later dispersed to their present locations. Which is the more likely scenario? Personally, I favor the hypothesis that C5-M356 has been introduced into South Asia as part of one of the hypothesized (post-)Neolithic migrations that has introduced a number of other haplogroups, such as R1a1a-M17, to the Subcontinent.
First of all J2 and R1a1 are not original from East Asia nor related to it in any meaningful way. So this you say could only make sense if C5 and C* would be original from West Asia or Northern/NW South Asia.
DeleteThen, looking at the matter of apportions in detail, we can clearly discard any correlation of C with R1a1.
However J2* (and only this lineage among those putatively foreigner to South Asia originally) is confusing. This may indeed indicate a massive inflow of farmers into the region, ultimately from West Asia and then have an impact similar to E-derived lineages had among the Hadza, Khoisan, Pygmy, etc. in Africa: inflitration by gradual immigration of neighbors (mostly males) and gradual incorporation of children born of relationships of tribal women with outsiders.
However the frequency of J2* among the HTF class is almost as high as among the "native" Tamil farmers (SC, DLF and AW classes) and higher than among the Brahmin-related class. Although it varies a lot by individual tribes. This seems to imply that, at some point in the past, J2* had much greater clout in the region than it has now (maybe along with H1*?)
As a passing note, I'd even consider that J2* might be a Paleolithic backflow from West Asia. After all the IJ and J nodes must be IMO from c. 40 Ka ago and there is no reason why some "Westerners" with J* and G (for example) could not have marched to South Asia, maybe in the context of the increased climatic pressure of the LGM (when it could have acted as "refuge" for peoples from Central Asia and Iran maybe). Just speculating but it's not impossible and better understanding of these clades in all the subcontinent is necessary before this can be wholly discarded either.
In any case, we agree that the root node of C must have been in SE Asia, right?
Then you are telling me a story (without any evidence I can see) of a subclade of C that includes C1, C3, C5 and C*-India. Where do you get that from? I never heard of it before but seems central in your discourse.
And, assuming you're right, why could not those lineages imply direct migrations between NE Asia and South Asia and not (as seems logical to me) a common origin in the intermediate region of SE Asia, also known in part as Indochina - for a reason (i.e. the region between India and China).
For me, the logical thing is that the common ancestor of all C subclades coalesced in the geographical midpoint of where the various subclades are found now (or not too far from it - it's not rocket science, you know). That centroid is clearly in SE Asia no matter how you look at it.
Even if you exclude C2, C4 and the Australasian C* (C6 or whatever), as you seem eager to do with no reason I can see anywhere, the midpoint between Southern India and NE China is still in South China (which is also part of SE Asia, depending who you read), let me guess without a map present: near Yunnan, right?
But first of all I have to see why on the Holy Phylogeny you associate the Asian subclades of C to the exclusion of the Australasian ones and only them. So far I know nothing about that except your word.
In the middle of the previous comment I forgot to insert a key remark, something like:
DeleteC is from SE Asia and J2* is from West Asia, how can they both be connected at all? More so without other lineages like R1a or O etc. being involved.
That's critically important. Together with the issue of the coalescence region of early C.
I have the feeling that some people from the Northern latitudes are secretly obsessed with bringing the center of history and prehistory to such latitudes, what is totally inconsistent with our basic tropical adaptations and very notably with the harsh realities of the Ice Age. Siberia and related regions were clearly very marginal in the Paleolithic. They did play a role in relation with NE Europe and, of course, Native America but that's about it. And definitely they were not involved in the major human flows that shaped population geography and genetics at least before the Neolithic (and really only limitedly after that).
"In understand that this finding of C5 together with C* in the southernmost regions of South Asia, with strong presence among foragers and together with other archaic-looking clades like F* and H* (and unlinked to O, which is the main marker of East Asian Neolithic influence) really brings the matter again to an scenario of very ancient South-SE Asia flows and backflows, what happened necessarily before c. 50 Ka., whatever the 'molecular clock' speculations say".
ReplyDeleteHere you go again with your Garden of Eden belief. There is absolutely no reason why all those Y-DNAs should have moved together from a single region. It is almost certain that they represent several distinct movements into Southern India. And, once more, molecular clock speculations have nothing whatsoever to do with things. It is just that you seem uncomfortable with the conclusions and are conjuring up anything at all that might justify your dismissing them. As Ebizur said:
"In any case, we agree that the root node of C must have been in SE Asia, right?"
I certainly don't agree, and I doubt that Ebizur does either. He wrote:
"In regard to the ancestry of C5-M356, it has been known since at least ten years ago (Redd et al. 2002) that, in median-joining networks of Y-STR variation, the majority of C-M130 representatives from the Indian Subcontinent cluster together and are well separated from Oceanic C2-M38, which seems to be the outlier among extant subclades of C-M130. Both Redd et al. and a later similar analysis by Hammer et al. have shown South Asian C5-M356 to be 'intermediate' between Japanese C1-M8 and East Asian/American C3-M217 in regard to Y-STR variation, yet each of these clades clusters separately despite being obviously more closely related to one another than any is related to Oceanic C2-M38"
So C5 is most closely related to the northern Y-DNA C lineages. That is reasonable evidence in favour of a Central Asian route east for the basal haplogroup. He also mentioned:
"C5-M356 is also relatively close to Australian C4-M210, although the latter clade deviates slightly in the direction of the very distinctive C2-M38".
That is not inconsistent with a Central Asia route to Australia either. C2 looks to have spent a great deal of time in the small islands of Southern Wallacea. That would lead to diversity of the haplogroup at one level and a great deal of drift at another, leading to the apparent paradox of a relationship between C4 and both C5 and C2.
Ebizur again:
ReplyDelete"As for South Asian C*-M130 Y-DNA, it seems to form a loose cluster with Southeast Asian C*-M130 Y-DNA, and this cluster of Southeast/South Asian C*-M130 seems to have diverged from the C-root around the same time as (and from an ancestor closely related to the ancestor of) Oceanic C2-M38".
So C* in India is almost certainly the product of movement from SE Asia. It is not indigenous to India.
"First of all J2 and R1a1 are not original from East Asia nor related to it in any meaningful way. So this you say could only make sense if C5 and C* would be original from West Asia or Northern/NW South Asia".
Not so. Ebizur has explained that the two C 'haplogroups' have separate origins. My guess is that C* entered India from the east while C5 entered India from the northwest. Ebizur again:
"I do not see any reason to presume that C5-M356 has diverged from a common ancestor with South/Southeast Asian C*-M130 within the Indian Subcontinent".
And neither do I. By the way, thanks for all the information concerning C. I was unaware of much of it.
"Then you are telling me a story (without any evidence I can see) of a subclade of C that includes C1, C3, C5 and C*-India"
No. Ebizur is telling you that C1, C3 and C5 are more closely related to each other than to any other C Y-DNA, and basically form a single clade within C. C* is not a member of that clade. The three haplogroups are next most closely related to C4, and finally only distantly related to C2 and C*.
"Where do you get that from? I never heard of it before but seems central in your discourse".
Ebizur told us in his second comment:
"Both Redd et al. and a later similar analysis by Hammer et al".
I suggest you read his comments again.
"C is from SE Asia and J2* is from West Asia, how can they both be connected at all?"
The trouble with that comment is you have constructed it on an incorrect hypothesis. C is probably not from SE Asia, just two clades, C2 and C*, are. Even these haplogroups must have come originally from somewhere near Africa.
"For me, the logical thing is that the common ancestor of all C subclades coalesced in the geographical midpoint of where the various subclades are found now (or not too far from it - it's not rocket science, you know). That centroid is clearly in SE Asia no matter how you look at it".
And you have told me meany times that centroids are unreliable as evidence for much at all. But, of course, in this example the use of a centroid suits your belief.
Sorry. Ebizur didn't say, 'In any case, we agree that the root node of C must have been in SE Asia, right?'. It was Maju. That come from shuffling my reply around. Sorry, again.
ReplyDelete"Ebizur told us in his second comment:
ReplyDelete"Both Redd et al. and a later similar analysis by Hammer et al"."
Thanks, I missed that. He actually said:
... it has been known since at least ten years ago (Redd et al. 2002) that, in median-joining networks of Y-STR variation, the majority of C-M130 representatives from the Indian Subcontinent cluster together and are well separated from Oceanic C2-M38, which seems to be the outlier among extant subclades of C-M130. Both Redd et al. and a later similar analysis by Hammer et al. have shown South Asian C5-M356 to be "intermediate" between Japanese C1-M8 and East Asian/American C3-M217 in regard to Y-STR variation, yet each of these clades clusters separately despite being obviously more closely related to one another than any is related to Oceanic C2-M38. C5-M356 is also relatively close to Australian C4-M210, although the latter clade deviates slightly in the direction of the very distinctive C2-M38.
The problem is that this is NJ-based on the evaluation of some STR markers, which are more than just often misleading.
If we ignore this issue and bluntly assume that the NJ tree is telling us the facts of the yet-to-be-unraveled phylogeny, then I'd like to know which clades are closer to the root (which can be tentatively identified by linking this haplotype tree with one of F, for example).
Anyhow, links are cool: I introduced "Hammer et al." on an scientific-specialized search engine and I come with a number of papers... on Jews, on human autosomal genetics...
After a lengthy journey of search, I arrived to an apparently relevant paper (Hammer 2005b), where in fig. 4d we get an NJ tree of haplotypes of C(xC2,C3) - careful there because it describes the tree as "C*" (which should be from context C(xC1,C2,C3)) but then it mentions that Japanese C1-M8 is included.
This C*+C1 only nature (C4 included but not C2 nor C3) can be confirmed by comparing circle size with apportions in fig. 2 but it should be clear from the legend also. No C2 and no C3 are shown in that graph.
Interestingly, as I foresaw just a few lines above this (when I still had not found the paper), South Asian and SE Asian lineages are intermediate, central, in the NJ tree, with the South Asian ones being very diverse (SE Asian ones not so much instead). Instead Japanese (C1) and most Oceanian (C4, not yet discovered but rather obvious as it forms a cluster) are peripheral to "true" C* (which also includes C5).
"True" C*, or more exactly C(xC1,C2,C3,C4) forms a cluster in SE Asia, which may well be a clade upstream of C4 (but still to be confirmed). However there are some Oceanian and SE Asian C* which remain in the central constellation, which is mostly South Asian.
This figure actually suggests very strongly that C coalesced in South Asia, deriving as follows:
→ C1 in Japan
→ C* SEA → C4 (= C* OCE)
→ C* small SA cluster (C5?)
→ many "private" (small) C* lineages in South Asia and (to lesser extent) SE Asia and Oceania
The exact position of C2 and C3 is not obvious from this graph.
I'm pretty sure now that Ebizur misunderstood this graph, because C2 and C3 are not displayed at all.
"... your Garden of Eden belief"...
I try to narrate in human words what the data says in raw facts.
Anyhow, how's my "tropical Garden of Eden" worse than your "frozen Black Hole" model? We are a tropical species and tend to live in or near the tropics if possible, more so when we lack the knowledge to survive in cold latitudes or the adaptations to low solar radiation needed for our vit. D photosythesis, so important for our brains.
I have included Hammer's NJ tree (annotated by me) as an update/appendix after the entry's main corpus, with my annotations. Take a look, please because it illustrates the answer to misunderstandings that seem to have fueled Ebizur's stand.
ReplyDeleteOf course, please do correct me if wrong, but I understand that the NJ tree actually suggests a South Asian origin for all C or at least the paragroup depicted there. That should explain why Japanese C1 looks as derived from South Asian C*.
It certainly looks more consistent with a South Asian rather than a SE Asian origin for C. However C4 does look like having a SE Asian C* precursor, something we cannot say of C1 (and we can't know for C2 and C3). Interesting.
I also included now the graph from Redd 2002. Everything in those graphs conspires to declare C original from South Asia and it is impossible to declare South Asian C "immigrant".
ReplyDeleteThe graph from Redd et al. 2002 is the one to which I have referred in my previous comments. The green "South Asian" circles at the top of the graph, between C1-M8 and C3-M217, are clearly C5-M356, as shown by the presence in this cluster of the white circle, which, as I recall, has explicitly been identified as an individual from Central Asia (Uyghur?).
ReplyDeleteThe other green circles toward the bottom of the graph represent South Asian C-M130 Y-chromosomes that are not closely related to the majority of South Asian C-M130 Y-chromosomes (C5-M356) and that seem to belong to a loose cluster of Southeast Asian/East Indonesian Y-chromosomes (along with an Oceanic and an Australian outlier). This whole cluster seems to be more closely related to Australian C4-M210 and Oceanic C2-M38 than to C1-M8, C3-M217, or C5-M356.
Of course I can only speculate at this point, but it seems likely to me that Japanese C1-M8, East Asian/American C3-M217, and South/Central/West Asian C5-M356 will eventually be found to comprise a subclade of C-M130 to the exclusion of at least Oceanic C2-M38 and Southeast/South Asian C*-M130.
I don't know how can you see much of what you say in those graphs (it looks terribly biased to me what you say, although some details are obvious and we do agree on them). Let's see:
Delete1. There are a few C* from India (two or four, depending on how loosely you consider that cluster) that approach the mysterious SE Asian cluster (which also includes two East Indonesians and one Oceanian). This cluster is at the bottom of C* in the Redd graph and (probably the same) between the (mostly Indian) core and the Oceanian cluster (which I read as C4) in the Hammer graph.
2. There is the probable C5 cluster above in the C* cluster in the Redd graph, not obvious in the Hammer graph.
3. There is a diverse array of Indian and Australian C* near the root in the Redd graph but from the Hammer one we see the Australian C* forming a distinct cluster which we know is C4.
4. So only Indian C* (and a few close relatives from SE Asia and Oceania) are definitely around the root of C (and quite diverse in there).
All this suggests, as did in fact to other researchers, that C coalesced in South Asia and quickly expanded to SE Asia and surroundings, which is the firs "coastal migration" model, as I remember it, which may have been outlined in 2006 or 2007 and was heavily based on this apparent structure of haplogroup C. I can't recall the exact paper but the model surfaced in those years because I remember discussing it and quickly burying (along with others) the pop-science Genographic "continental" model for the OoA and Eurasian expansion, which also failed to explain many things about other lineages (F, D, etc.)
... "but it seems likely to me that Japanese C1-M8, East Asian/American C3-M217, and South/Central/West Asian C5-M356 will eventually be found to comprise a subclade of C-M130 to the exclusion of at least Oceanic C2-M38 and Southeast/South Asian C*-M130".
It might be because in Y-DNA all nodes are two-branched (if we'd know all the actual SNPs) because every generation introduces new SNPs almost necessarily. But the connection is tenuous so it'd be a rather unimportant node in the chain from C-root to the known major subclades anyhow.
Still the haplotype NJ trees are not nor can be informative enough on their own, as we can see when we compare the two trees, in which the same clusters (or very similar ones) organize very differently - very illustrative of this problem Australian Aboriginal presumed C4, which in one tree appears disorganized and near the root and in the other organized at an extreme.
So I trust the SNP-based phylogeny and these only as suggestive but blurry complement.
"I don't know how can you see much of what you say in those graphs"
ReplyDeleteI've had time to look at them and I can actually see what Ebizur is getting at. Although both are fairly old papers that doesn't make them unreliable. But we have to remain aware of what the authors were looking for.
Firstly the Hammer paper. The author was specifically concerned with C1 and its relationship to a select few Y-DNA Cs. I think you are mistaken in labeling the blue dots C4. The two colours are far more likely to represent C2 and C2a as they are given as 'SE Asia' and 'Oceania', not 'Australia'. Therefore C3 and C4 are not included at all. So we are left with a distant connection between C1 and some South Asian haplogroups, probably both C* and C5.
Secondly, the Redd paper. As you say, it is even more interesting than the other paper. Your comment:
"While C4 is not obvious here, the fact that South Asian (Indian subcontinent) C* is central to all the haplogroup is again underlined".
Taken at face value it is actually impossible to claim either South Asia or Australia as being 'central to all the haplogroup'. I agree that 'Australia' is unlikely, but what has happened since this paper came out is that Australian C has been designated 'C4'. That means that the 'Australian Aboriginal' C in the diagram should be all C4, whereas it is shown mixed with South Indian C (C*?).
"but from the Hammer one we see the Australian C* forming a distinct cluster which we know is C4".
I see no 'distinct cluster' of the Aboriginal Australian haplogroups at all. Several Indian haplogroups are scattered through the Aboriginal Australian ones. That is unlikely to be the case in fact. Remember that the authors' objective at the time was to find evidence for the then becoming popular great southern coastal migration. Therefore the fact that the connection between India and Australia has become very blurred in the diagram is not too surprising. So we have to tease out the Australian Y-DNA C from the Indian. And the RPS4Y/M216 point of connection in the diagram is likely to be unreliable.
I agree that 'The protuberance to the top might be C5', in fact almost certainly is. As Ebizur wrote, 'The green "South Asian" circles at the top of the graph, between C1-M8 and C3-M217, are clearly C5-M356'.
"All this suggests, as did in fact to other researchers, that C coalesced in South Asia"
Possibly so. But once we consider that to be the case, and we place South Asia as the origin for the haplogroup, a whole new possible sequence of mutation is revealed. We have C5 as the earliest branch or 'origin', followed by C3 (top right). C1 is next (top left) and then we have the mess that is C4 mixed with C*, a mix of Aboriginal Australian with various C* in SE Asia and India. This Indian C*, though, is not part of the C5 clade. It is separate. C2 then apparently branches off this C*, but this part of the diagram obviously needs a closer examination before any real conclusions can be drawn.
The Hammer graphic is indeed focused on C1, which is the only clade mentioned in the legend other than C*, which is the title of the graph. In the previous table, C* is clearly described as C(xC1,C2,C3) because C4 was not yet described as such. So the graphic is one of C(xC2,C3) with C1 marked as distinct by the transversal bar.
DeleteThe distribution mentioned in the previous table for C(xC1,C2,C3) fits almost ideally with what we see in the equivalent part of that image labeled, let's not forget, C* and not C.
"Taken at face value it is actually impossible to claim either South Asia or Australia as being 'central to all the haplogroup'".
By that graph indeed. But we also have the other graph to complement our understanding and very few Oceanian lineages are at the center there.
As I said before, don't try to get a perfect resolution from haplotype graphs: they are sometimes informative but sometimes confusing. This may be partly dependent on the number and relevance of the STR markers considered but in any case it is a well known fact that haplotype coincidence, especially between haplogroups is only suggestive and not proof of anything.
Someone check this but it's likely that Hammer used more markers (while Redd was more ambitious in the sampling and analysis instead).
"Remember that the authors' objective at the time was to find evidence for the then becoming popular great southern coastal migration".
Redd 2002 is previous by many years to the "revolutionary" proposition by Macaulay 2005 of what they described as: "single, rapid coastal settlement of Asia". Hammer 2006 (prepared in 2005) overlaps with Macaulay's key paper.
Regarding the last you say, I would not try to read too much in those graphs, notably not a chronology. If I'd do anyhow, I would see a quasi-starlike structure with at least five major branches: C1, C2, C3, the possible C5 cluster and the SE Asia centered cluster.
If we see it as a starlike structure, then it'd be even more clear signature of a rapid expansion. But, then again, I don't see how can we infer too much from these old haplotype graphs. We'd need at least a better STR marker pre-selection, rendering a more neat graph.
Thanks for that reasoned reply. I agree that more work on the haplogroup is needed before we can come to any realistic conclusions. But regarding:
ReplyDelete"Redd 2002 is previous by many years to the 'revolutionary' proposition by Macaulay 2005 of what they described as: 'single, rapid coastal settlement of Asia'".
I was certainly well aware of the proposition some years before 2005. I saw a program on pay TV by Spencer Wells claiming such a migration. I had pay TV only for a couple of years around 2002. Here is a Wiki link about the program:
http://en.wikipedia.org/wiki/The_Journey_of_Man:_A_Genetic_Odyssey
I could immediately see the huge flaws in his argument for a specifically 'coastal' migration through South Asia to Australia. For a start the close connection he claimed to see between Indian and Australian haplogroups was very difficult for anyone else to see. Just the Y-DNA C in common, and not even a particularly close relationship within that haplogroup which is spread through Central Asia as well. Here's an extract from the film that you may find interesting:
http://studgen.blogspot.co.nz/2007/07/spencer-wells-in-australia-and-india-5.html
Well, I'm sorry not to be familiar with the pop-science of that Wells guy (I say pop-science because where is the paper?, after all).
DeleteIn any case, I do agree that the coastal migration model in its strict interpretation is probably not right but I stand for a more flexible variant: the tropical migration model (coastal, riverine or merely by regular walking), which fits way too well with both the genetic and archaeological evidence not to be correct.
The Redd (2002) paper is actually older than Wells' TV program so the idea was obviously around at the time.
ReplyDelete"Well, I'm sorry not to be familiar with the pop-science of that Wells guy (I say pop-science because where is the paper?, after all)".
Exactly. Pop science it is. In fact the idea infiltrated pop science before it became the subject of scientific research. As a consequence the scientific research started out with a particular objective: to find evidence in favour of the Great Southern Coastal Migration. They put the cart before the horse, as some have it. From that point on the evidence has consistently been interpreted with that particular objective in mind. Each individual piece of inconvenient evidence uncovered during the research that might lead to some other conclusion has been immediately explained away by invoking drift, bottlenecks or founder effect. That's really convenient, because the phenomena can be invoked or not, depending on what the researcher wishes to find. That's why I compare drift, bottlenecks or founder effect to the expression 'abracadabra'. They can make things disappear whenever convenient. The Redd paper is even an example of the putting of the cart before the horse. The author specifically sets out to 'prove' a connection between India and Australia, no matter how tenuous. The author even finishes up with an unlikely Holocene connection:
"Phylogenetic analyses of STR variation associated with a major Australian SNP lineage indicated tight clustering with southern Indian/Sri Lankan Y chromosomes. Estimates of the divergence time for these Indian and Australian chromosomes overlap with important changes in the archaeological and linguistic records in Australia. These results provide strong evidence for an influx of Y chromosomes from the Indian subcontinent to Australia that may have occurred during the Holocene".
Those 'changes' actually fit far better with the expansion of Y-DNA K than with C anyway.
I thought you were a fan of Wells, haha!
DeleteWhatever the case it's not putting the cart before the horses because the method is correct: someone(s) outline a hypothesis and they or others try to find evidence supporting (or rejecting) it: that's the scientific method in fact:
0. Previously known data
1. Hypothesis drafting
2. Evidence search
3. Theory formation
4. Scientific controversy: evidence contrasting, experiment replication, finding of new data
5. Theory consolidation
... etc.
A pop hypothesis is as good as any, as long as it makes sense and can be contrasted. But I never heard of the tropical route before 2005 or 2006, maybe even 2007. The mainstream model was another pop-science conjecture, which was being promoted (on no evidence, just hunches) by the Genographic Project.
Some geneticists had hunched the Siberian route, I guess, based on the relative rarity of N and R in South Asia and the generally accepted preconception that West Asia was the region of radiation of Homo sapiens after the exit from Africa. This model was kind of "self-evident" (on first sight) but in the end it did not fit well with the data. As we kept learning more and more from mtDNA and Y-DNA almost parallel phylogenies, we had to accept that West Asia was not the source of the Eurasian+ radiation but that this one was in Southern Asia (South and SE Asia essentially). We could not find evidence of Homo sapiens in Siberia before Australasia and Southern Asia and in the end people made facepalm on the matter: of course!, why would we go with our tropically adapted nakedness to the cold reaches of Siberia if we could equally (or easier in fact) go to the temperate tropics of Southern Asia first? Facepalm!
It should be no more problem and readily acceptable to all but almost one or two centuries of Nordicism had made an impact in our mentalities and made some blind to the obviousness and stubbornly so. The same kind of people who were ready to accept Piltdown Man and reject the true African ancestry of Humankind.
...
...
Delete"Each individual piece of inconvenient evidence uncovered during the research that might lead to some other conclusion has been immediately explained away by invoking drift, bottlenecks or founder effect".
LOL. I'd dare say that it's you, like the infamous Russian anthropologist who claims even to this day an American origin of our species against absolutely ALL the evidence, who need bottlenecks all the time to "explain" the necessary extinction of every single piece of genetic evidence from Siberia and nearby areas. For you "evidence" is like a river and is long gone before we arrived, so you can imagine whatever fits your fantasy and claim that X people was in Y place but that they moved or were displaced/massacred long ago, because in your mind that's what people do: walking forth and back through Siberia and never remain attached to any place or region.
Maybe it's just that as white New Zealander you lack deep roots and imagine everyone being the same but whatever the reason it is just a narrative you make on zero evidence because YOU appeal to permanent mobility and massive imagined bottlenecks to "explain" why the evidence isn't there as it should.
Luckily archaeology does not support your nonsense while it does support more and more the tropical migration model.
Argh, so tiresome! Again with this? When are you going to retire, Terry?!
"The Redd paper is even an example of the putting of the cart before the horse. The author specifically sets out to 'prove' a connection between India and Australia, no matter how tenuous".
While he's wrong you are being unfairly merciless with him: his data suggested that (and there were pre-existent anthropometric theories favoring that idea: the infamous "Australoid" type and all that). And that's the good thing of reading it 10 years after publication (and what 10 years of continuous discovery!), that we can spot the flaws and limitations - and yet his graph, his raw data, is still largely valid and somewhat informative.
"A pop hypothesis is as good as any"
ReplyDeleteFor you, obviously.
"It should be no more problem and readily acceptable to all but almost one or two centuries of Nordicism had made an impact in our mentalities"
It may have impacted your mentality but it certainly has no influence whatsoever on mine.
"Some geneticists had hunched the Siberian route, I guess, based on the relative rarity of N and R in South Asia"
And those haplogroups don't fit the 'tropical route'. The idea that there was only a single route east and north from Africa is a ridiculous position to take. It springs solely from the Garden of Eden hypothesis.
"who need bottlenecks all the time to 'explain' the necessary extinction of every single piece of genetic evidence from Siberia and nearby areas".
And you are forced to call on drift, bottlenecks and founder effect to 'explain' 'the relative rarity of N and R in South Asia'. What's the difference between you and 'the infamous Russian anthropologist'?
"why would we go with our tropically adapted nakedness to the cold reaches of Siberia if we could equally (or easier in fact) go to the temperate tropics of Southern Asia first?"
Our 'tropically adapted nakedness' did not prevent pre-sapiens Homo species from entering northern Anatolia or even North China. Surely if those species could do so it is inconceivable that H. Sapiens was unable to do so. And I would suggest that entry to South Asia is not so easy. There is very little evidence of haplogroup exchange between SW and S Asia. And exit from India to the east through jungle-covered mountains and the Ganges-Brahmaputra Delta is even more difficult. Compared to that a movement through regions of Siberia is a doddle.
"We could not find evidence of Homo sapiens in Siberia before Australasia and Southern Asia"
You know as well as I do that absence of evidence is not evidence of absence.
"For you 'evidence' is like a river and is long gone before we arrived, so you can imagine whatever fits your fantasy and claim that X people was in Y place but that they moved or were displaced/massacred long ago"
I have never ever made any such claim. I merely look at the evidence as it stands. No displacement or genocide involved. Where an apparently early haplogroup now makes up a small proportion of haplogroups I suggest it is because it became outnumbered by the arrival of later ones.
"YOU appeal to permanent mobility and massive imagined bottlenecks to 'explain' why the evidence isn't there as it should".
I defy you to come up with a single instance where I have done such a thing.
"And those haplogroups [mtDNA N and R] don't fit the 'tropical route'".
DeleteActually they do but it's not as simple as looking at frequencies: basal diversity must be considered also.
"The idea that there was only a single route east and north from Africa is a ridiculous position to take".
No idea what this means. Excluding Europe (which we know was excluded for some 70 Ka maybe), the only route out of Africa is necessarily in NW direction and ends up in Iran, surely in the swampy Persian Gulf Oasis (as further North was Neanderthal territory). The debate should not therefore be not about the OoA but about the Out of the Persian Gulf.
Your propositions are often wrong and made out of 90% anger with a thin shell of pseudo-reasoning. Why so much anger? Why not to accept that others may be right after all?
... "you are forced to call on drift, bottlenecks and founder effect to 'explain' 'the relative rarity of N and R in South Asia"...
Not more than would be needed for your Siberian model, actually less. R is not "rare" in South Asia at all (and I contend that it coalesced in South Asia in fact) but N does look original from SE Asia (or is that Bengal?) what implies some sort of long stem since Africa ("bottleneck"? not my choice of words certainly because there's no previous "bottle" to be "necked" to begin with.
Whatever the case we are faced with a pre-N migrating from Africa to SE Asia in "seed" form (i.e. without expansions that left any mark) either in the tropical route or in the subarctic route models. But the tropical route is much shorter and easier and, guess what?, it can be sped up by means of boating (at least some argue that).
So I invoke here (again) goddess Parsimony and her magical weapon: Occam's Razor!
"Our 'tropically adapted nakedness' did not prevent pre-sapiens Homo species from entering northern Anatolia or even North China".
It probably did to some extent. Those sites are relatively late arrivals: much later than the Eurasian expansion in the tropics with all likelihood. However they are still mild, almost "warm", compared with the hellish Siberian winters.
"And exit from India to the east through jungle-covered mountains and the Ganges-Brahmaputra Delta is even more difficult".
That's a knee jerk dogma you have proclaimed and is actually false. You don't even know the climate of those hills in the Ice Age (surely drier and fresher) but in any case those muddy jungles are no true barrier, just a very much passable obstacle.
... [don't go yet, the best laughs come now]
...
Delete"Compared to that a movement through regions of Siberia is a doddle".
ROFLMAO
I can just visualize a semi-naked Terry wading through belly-high snow, his long hair covered in frost in his last survival effort, his very last seconds of life and claiming defiantly: "just a doodle!"
Altai freezes in Winter even today, mind you. It's not easy to inhabit: learn first, babble later.
Barnaul is the capital of Altai Krai, where Paleolithic hominins of several types inhabited. It's climate is most interesting for our debate:
The continental climate of Barnaul (Köppen Dfb) is defined by its geographical position at the southern end of the Siberian steppe: it is subject to long, frigid winters, with an average of −17.5 °C (0.5 °F) in January...
It's not just January as you can probably imagine by now but half the year: the average high is below 0ºC (freezing point) between December and March and the average low is below 0ºC between October and April. Only a very short summer is really alright.
And we are not discussing present Interglacial (warm) climate but the Ice Age! To live in Altai, and therefore to be able to migrate through the Siberian route, you need very good specialized technology (and/or biology) and cultural memes... unless you take the Transiberian Railway.
It should be slashed out as soon as we consider it. (More so considering that any evidence of Homo sapiens in the area is more recent than anywhere else that matters, except America).
"You know as well as I do that absence of evidence is not evidence of absence".
Hmmm... I'm glad that you finally accept that. Still absence of evidence is not evidence either (facepalm!) and you never present any evidence whatsoever.
"I merely look at the evidence as it stands. No displacement or genocide involved. Where an apparently early haplogroup now makes up a small proportion of haplogroups I suggest it is because it became outnumbered by the arrival of later ones".
ROFLMAO again. So they were outnumbered but there was no displacement nor genocide, not even an accidental one...
And what about the massive loss of diversity. If Eurasian haplogroups of any type would have stem from Altai, we should see at least a remnant of that basal diversity. We see nothing of that but a highly derived and subset lineages with very low diversity in fact.
No bottleneck? No extinction? No genocide? No, of course not because, to begin with, the Eurasian macro-haplogroups did not stem from there nor went through there. Only some derived branches did at a very late stage of the Eurasian colonization in fact.
"I defy you to come up with a single instance where I have done such a thing".
You just did two quotes above. Your denials are confessions.
"I can just visualize a semi-naked Terry wading through belly-high snow, his long hair covered in frost in his last survival effort, his very last seconds of life and claiming defiantly: 'just a doodle!'"
ReplyDeleteI'm sure you will be able to explain exactly how pre-sapiens humans survived in Central Siberia while sapiens did not have the intelligence to do so.
"Why so much anger?"
I have no anger whatsoever, Maju. In fact I find trying to explain things to you rather amusing. Your prejudices and contradictions are absolutely astounding.
"Not more than would be needed for your Siberian model, actually less"
Maju, if you're going to claim a SE Origin for any haplogroup you are forced to postulate a genocide for through India for that haplogroup. A route through Central Siberia for several haplogroups requires no genocide whatsoever. And we know that climate change led to humans disappearing and reappearing several times in the region that became Britain. Surely you are able to accept the same phenomenon for other regions of Eurasia.
"R is not "rare" in South Asia at all (and I contend that it coalesced in South Asia in fact)"
That's a ridiculous statement seeing that you have just said:
" it's not as simple as looking at frequencies: basal diversity must be considered also".
R's greatest basal diversity is in SE Asia by a long shot.
"but N does look original from SE Asia (or is that Bengal?) what implies some sort of long stem since Africa"
And a genocide through the whole region between Africa and Bengal.
"Whatever the case we are faced with a pre-N migrating from Africa to SE Asia in "seed" form (i.e. without expansions that left any mark) either in the tropical route or in the subarctic route models".
That is a far more complicated (and unlikely) scenario than any I have postulated.
"So I invoke here (again) goddess Parsimony and her magical weapon: Occam's Razor!"
You're not invoking Occam's Razor at all. You're making things up!
"It probably did to some extent. Those sites are relatively late arrivals'
No they're not. You're making things up again to suit your belief. Humans have certainly been in Northern China since a million years ago for a start.
"That's a knee jerk dogma you have proclaimed and is actually false. You don't even know the climate of those hills in the Ice Age (surely drier and fresher) but in any case those muddy jungles are no true barrier, just a very much passable obstacle".
That statement is absolute rubbish. I have drawn your attention to the steep cline between Y-DNA O/Mongoloid and the Papuan phenotype in Indonesia. We see a similar steep cline in the Himalaya region between Y-DNA O/Mongoloid and South Asian phenotype. In both cases the Mongoloid/Y-DNA O has spread beyond the geographic boundary, but only a little. Although the Y-DNA O/Mongoloid and South Asian phenotype is not as steep in Northeast India it is still very steep. The route between Northeast India and SE Asia is almost as marked a geogrphic boundary as are the Himalayas and Wallace's Line. On the other hand the Mongoloid/Y-DNA O cline in Central Asia is very gradual. That supports a more navigable geographic boundary in that region although perhaps only going back as far as the Bronze Age, but possibly of very ancient standing.
"R's greatest basal diversity is in SE Asia by a long shot".
DeleteIn your maddest dreams maybe! Can you even count before making me waste my time again?!
West: 11/18 lineages (South Asia: 9/18 = 50%)
→ South Asia exclusive: R5, R6, R7, R8, R30, R31
→ shared SA+WEA: R1, R2'JT, U
→ WEA only: R0, R3
East: 7/18 (SE Asia: 5/18 = 28%)
→ SE Asia: R9 (incl. F), R11'24'B, R12'21, R22, R23
→ Australasia: R14, P
Even if we discard U, because it does look like a back-migrant into South Asia, the subcontinent still holds a clear majority. Even if you go Terry-style nit-picky and count only those lineages found exclusively in South Asia, they are still more than those found in SE Asia altogether (6 vs 5).
Count, dammit, count!
"And a genocide through the whole region between Africa and Bengal".
Not really: N just never found a niche to expand in. Alternatively you can blame Toba ash... but whatever: the case is that a Siberian route model is not better either.
"I'm sure you will be able to explain exactly how pre-sapiens humans survived in Central Siberia while sapiens did not have the intelligence to do so".
Sapiens did... in due time. I can't be sure of the details (thick natural furs or artificial means) but in any case those species had been living in the region (Eurasia, including maybe high latitudes) for maybe more than one million years, so they had time to adapt, regardless of whether this adaption was cultural or biological.
"You're making things up!"
What am I making up?
I'm bored of falling into your honey traps of false reasoning anyhow, so time again to agree to disagree I guess.
"Altai freezes in Winter even today, mind you".
ReplyDeleteNo doubt you will find this interesting then:
http://archaeology.about.com/od/dathroughdeterms/qt/denisova_cave.htm
Quote:
"Located in the northwestern Altai Mountains some 6 km from the village of Chernyi Anui, the site shows human occupation from the Middle Paleolithic to the Late Middle Ages, beginning ~125,000 years ago".
And this:
http://link.springer.com/article/10.1007%2FBF00974881?LI=true
Quote:
"Considerable progress has been made in recent years in the study of the Lower and Middle Paleolithic of northern Asia. There is growing evidence for initial human occupation before 700,000 years ago—as early as elsewhere in Asia—and for a very early adaptation to the arctic desert environment. New models of Lower Paleolithic settlement involve expansion and reduction of occupation in response to climatic variation, rather than simple colonization followed by steady occupation. The Middle Paleolithic of northern Asia is better documented, including actual finds of archaicHomo sapiens. The transition to the Upper Paleolithic seems to involve the survival of earlier cultural traits"
How do you propose to explain that?
"don't go yet, the best laughs come now"
No, they come now, when you attempt to explain the presence of pre-sapiens humans in Central Siberia.
"How do you propose to explain that?"
DeleteThick fur? Probably the correct answer. Maybe it's Denisovans what tickled down the collective imaginary into the idea of the Yeti? Why not?
Sorry:
ReplyDelete"and yet his graph, his raw data, is still largely valid and somewhat informative".
Except for the most striking thing about his diagram: the obvious closer relationship between Y-DNA C5 and both C1 and C3, rather than with C2. Talk about manipulating the evidence so you can see only what you want to see! I look forward to further entertainment as you wriggle your way out of that too.
I don't see that. I see three branches hanging from a messy, ill-defined, center. You can't take haplotype NJ links as true phylogenies, they are just suggestive stuff outlining possible paths for further research.
DeleteDo you want to over-read in that? Your problem. Go find someone else that listens to your speculations.
Correction:
ReplyDelete"On the other hand the Mongoloid/Y-DNA O cline in Central Asia is very gradual".
The Mongoloid element of that cline is not just confined to Y-DNA O of course. It is supplemented with the northern version of NO: Y-DNA N. Y-DNA C3 also contributes to the Mongoloid element in the region also. In both South and Central Asia the Y-DNA has moved much further than the Mongoloid Phenotype though. O2 has reached Pakistan and N has reached Northern Europe.
"I don't see that. I see three branches hanging from a messy, ill-defined, center".
ReplyDeleteYou don't see it because you don't want to see it. I agree that further research is needed before meaningful conclusions can be made but you did say, 'and yet his graph, his raw data, is still largely valid and somewhat informative'. The only 'informative' aspect I can see is the connection between the three C haplogroups.
"Thick fur? Probably the correct answer".
What possible evidence do you have that Neanderthals or Denisovans had 'thick hair'? I would have thought that the human general lack of hair had developed in the tropics during Homo erectus times.
"In your maddest dreams maybe! Can you even count before making me waste my time again?!
West: 11/18 lineages (South Asia: 9/18 = 50%)
→ South Asia exclusive: R5, R6, R7, R8, R30, R31
→ shared SA+WEA: R1, R2'JT, U
→ WEA only: R0, R3
East: 7/18 (SE Asia: 5/18 = 28%)
→ SE Asia: R9 (incl. F), R11'24'B, R12'21, R22, R23
→ Australasia: R14, P"
Check again. Exclusive to South Asia we have the six haplogroups you mentioned. R1, R2'JT, U can hardly be considered 'South Asian'. R1 is hardly common there, R2'JT is basically only present (in very small numbers) as R2, and U is present basically only as U2 and U7 although these two comprise the most frequent R haplogroups in India. The three haplogroups you calim as 'shared' are, like U, most likely to be a back movement into South Asia from further west. As for your 'Australasian' haplogroups: what a joke. P is found in the Philippines, although almost certainly having an Australian origin, and R14 is found from New Guinea through the lesser Sunda Islands and west into the Nicobar islands. That makes it a SE Asian haplogroup. And R11'24'B is in effect three haplogroups united by a single control region mutation, a mutation you are quite prepared to ignore when it suits your purpose. That gives us 8 SE Asian haplogroups.
"Not really: N just never found a niche to expand in".
How on earth do you propose that happened? Surely India provides evidence that multiple haplogroups managed to survive, and thrive, there. But here you go claiming that poor old N couldn't gain a foothold. Get real. If it had 'never found a niche to expand in' it wouldn't have survived its journey through South Asia.
"I would have thought that the human general lack of hair had developed in the tropics during Homo erectus times".
DeleteOf course there's a lot of uncertainty on the matter but remember please that there was this study (Toups 2011), which proposed that:
1. For the Homo sapiens line Clothing emerged c. 200 Ka ago, what is within the the Homo sapiens range as distinct species.
2. Neanderthal ancestors, Homo heidelbergensis, made clothes or at least leatherworking c. 780 Ka ago. Neanderthals and their ancestors also used fire commonly, what suggests low or no fur coverage of the body.
3. Fur loss is estimated to be c. 1200 Ka ago, not long before the separation of the lines leading to Neanderthals and Homo sapiens, but long after the divergence of the Asian H. erectus branch.
Also Asian H. erectus and their Denisovans relatives are not related with fire remains as far as I know, what would seem to add weight for a fur coverage of their bodies.
Everything is quite tentative, uncertain, but this could well be a reasonable scenario. Also there's no reason not to imagine that Heidelbergensis/Neanderthals could have re-evolved at least in part some fur coverage considering their habitat for about a million years.
As for South Asia and R, you are just in denial: even counting your way, which is totally arbitrary, South Asia has the top count. Also you can't really consider Wallacea as SE Asia proper but as transitional between SEA and Sahul.
But whatever, cheat all you want, the weighted centroid falls in South Asia no matter what.
"How on earth do you propose that happened?"
Small private lineages are found often enough. Pre-N would not be the only one in such situation; for example R3 is one such tiny private lineage surviving as such in West Asia probably since c. 55 Ka ago or so. Why? Call it "oasis dynamics" or whatever.
Pre-N carriers may even have migrated to South Asia at a late stage of the colonization or whatever. Not only Pre-N did not survive anywhere but it did not survive in any of its many incarnations (five coding region mutations between L3 and N allow us to talk of four distinct pre-N variants which must have existed at some point in time and space, but do not anymore: L3n, L3n1, L3n1a, L3n1a1... and finally N itself).
IF L3n1a1 was carried by a boater-specialist population which followed the rapid coastal migration model strictly, then you have your explanation version A.
Another possible explanation is that their few descendants in South Asia were drifted out by the genocidal effects of Toba ash.
Finally it's possible that N actually expanded from South Asia in spite of not leaving many direct descendants there (only N1'5, N2 and R).
Whatever - but certainly not Siberia.
I remain unconvinced concerning the fur.
ReplyDelete"Small private lineages are found often enough. Pre-N would not be the only one in such situation; for example R3 is one such tiny private lineage surviving as such in West Asia probably since c. 55 Ka ago or so".
But R3 hasn't inexplicably moved across half the globe. It's route there is scattered with its relations.
"Pre-N carriers may even have migrated to South Asia at a late stage of the colonization or whatever".
With that reasoning you are able to make up anything you like.
"Also you can't really consider Wallacea as SE Asia proper but as transitional between SEA and Sahul".
And yet you can consider the whole India subcontinent as a single region? And talking of that transitional region: to me the most convincing evidence that R coalesced in SE Asia/Wallacea is the distribution of basal N haplogroups in the region. We have N13, N14, O and S in Australia, but nothing in that transitional region. The nearest other basal N haplogroups are N21 and N22 on mainland SE Asia, from Laos to the Malay peninsula. The only N-descended haplogroups through what became island SE Asia(Indonesia, the Philippines and island Malaysia) are R-derived. The most easterly R-derived haplogroup is P. To me it obviously originated in Australia with N13, N14, O and S. But eventually it was to spread along the edge of the Pacific Ocean from the Philippines in the north to Fiji in the east. R23 straddles Wallace's Line, R22 is present on mainland SE Asia but notably also on the Lesser Sunda Islands (almost certainly the route N took from mainland SE Asia to Australia), R12'21 splits into an Australian haplogroup (R12) and one that survives in Malay Negritos. R14 is spread from the Nicobar Islands to New Guinea. R9 is basically a mainland SE Asian haplogroup, although possibly originating in Sumatra. And further:
"As for South Asia and R, you are just in denial: even counting your way, which is totally arbitrary"
You are surely prepared to accept that R11'B6, B4'5 and R24 separated very soon after, and very near where, R first appeared. As a single haplogroup R11'24'B is spread in an arc from South China, mainland and island SE Asia to the Philippines.
"But R3 hasn't inexplicably moved across half the globe. It's route there is scattered with its relations".
DeleteBut, but, but... my butt. When I try to answer your rather absurd questions, you always manage to reach to some other burning nail.
Seriously: I simply do not see any evidence whatsoever of any migration of Homo sapiens through the Altai corridor prior to 40 Ka. or so. That includes all the "buts" you figure out for N squared (because the route through Siberia is doubly long and many times more difficult) but also includes archaeological evidence, which in Altai only points to archaic hominins and not Homo sapiens before the Aurignacian period.
Stop making questions to me and make questions to yourself. Reality rules in any case.
Now, to your complaint, there are of course many private lineages in all kind of situations but most people just choose to ignore them calling them whatever-asterisk. Also the closest relative of N is M (and the other L3) and when N arrived to SE Asia (or wherever it boomed) it did with M subclades quite obviously.
For each of your "buts" there is plausible explanation but before you ask again try to answer them yourself for the "Siberian route": in all cases there is not a single improvement with your model and in many there is serious loss of plausibility and parsimony.
"And yet you can consider the whole India subcontinent as a single region?"
Actually I tend to consider the Ice Age South as separated (ASI component) and melting with the North (ANI) mostly after the Ice Age only. But it does not matter too much.
Evidence of a different kind emerges if we are prepared to actually consider the possibility that the R haplogroups may have entered India from the east. R7 is notably present in Munda-speaking populations, a language possibly related to Austro-Asiatic. It is centred on Northeast India but has spread, especially along the East coast and through regions accessible from the Godavari River. R8 too looks perhaps to have originated on the east coast, this time north of the Godavari in Orissa.
ReplyDeleteThat leaves just 4 haplogroups that may provide unequivocal evidence that R originated in India, not in SE Asia. But, as well as being especially common in Kashmir, R6 is common along the southeastern coastline. But it has also been found in Central Indian tribal groups. Perhaps R6 too was part of an expansion through India from the eastern coastline. Perhaps it was part of the Great Southern Coastal Migration, but in the wrong direction.
So we're down to 3 haplogroups that might definitely support an India origin as opposed to a SE Asian one. R30 is especially common in those regions easily accessible from the Ganges River as far upstream as Rajasthan and the Punjab. The two remaining Indian R haplogroups may have been in the vanguard of a population moving via the rivers, from the east. After all, we know that several R haplogroups coalesced even further west beyond the Indian subcontinent. R5 is spread right across India but not towards the southeast. R31 is basically just further west than is R5.
Not true, Terry: the lineages are all South Asian (and not East Asian) in all cases and by no means restricted to minority populations like the Munda.
DeleteGo write your own blog and stop torturing me with your pointless nagging and wailing.
"Go write your own blog and stop torturing me with your pointless nagging and wailing".
ReplyDeleteI am trying to point out where your suppositions are completely wrong.
"Not true, Terry: the lineages are all South Asian (and not East Asian) in all cases and by no means restricted to minority populations like the Munda".
Maju, you know as well as I do that the Indian lineages are not evenly distributed through the subcontinent. To claim a South Asian origin for haplogroup R you would have to show which individual region within the subcontinent had the greatest basal diversity. And furthermore you would have to demonstrate that particular region had a greater diversity than does a region of similar size anywhere else. Specifically in this case, SE Asia.
"But, but, but... my butt. When I try to answer your rather absurd questions, you always manage to reach to some other burning nail".
Your comment about R3 had no relationship whatsoever to the matter in hand. It was a complete red herring proposed only to confuse the issue.
"Seriously: I simply do not see any evidence whatsoever of any migration of Homo sapiens through the Altai corridor prior to 40 Ka. or so".
That may be so. But there is ample evidence against mt-DNA N having moved east through India. So we are left trying to find an alternative route for the haplogroup.
"which in Altai only points to archaic hominins and not Homo sapiens before the Aurignacian period".
Are you now claiming the H. sapiens left Africa with a fully-formed Upper Paleolithic technology?
"Stop making questions to me and make questions to yourself. Reality rules in any case".
I'll answer the question myself then. Yes, you do believe what I just asked, but that is an extremely unlikely scenario. The Upper Paleolithic developed some time AFTER H. sapiens had emerged from Africa. What's more it developed outside Africa. That surely solves any problem you imagine about a lack of Upper Paleolithic in the Altai at any possible period for N's expansion.
"Now, to your complaint, there are of course many private lineages in all kind of situations but most people just choose to ignore them calling them whatever-asterisk".
And almost without exception it can be assumed that those private lineages first appeared very near where they are now found. Their 'long tail' developed in situ, not while they were traipsing across miles and miles of ideal habitat. In fact I can think of no exceptions to that scenario.
"Also the closest relative of N is M (and the other L3)"
M is no more closely related to M than it is to the other L3 haplogroups as far as I'm aware. It's expansion is therefore almost certainly independent of M's.
"when N arrived to SE Asia (or wherever it boomed) it did with M subclades quite obviously".
Not 'obviously' at all. Only in your imagination. If it had traversed India with M it surely would have been able to establish itself occasionally on that continent as well as M did. Unless you're yet again postulating genocide.
"For each of your 'buts' there is plausible explanation"
But every explanation you have made up is completely implausible.
"To claim a South Asian origin for haplogroup R you would have to show which individual region within the subcontinent had the greatest basal diversity".
DeleteI could do that, in fact I have done it in the past but you have not taken due note but rather stormed in anger or changed theme, learning nothing. Whatever the case your demands of evidence are so twistedly excessive that you could declare Hitler not guilty of genocide on a technicality.
"Your comment about R3 had no relationship whatsoever to the matter in hand. It was a complete red herring proposed only to confuse the issue".
On the contrary: we see many examples every day of very small private lineages who are alive and well, often have been for tens of thousands of years. We also see long migrations like pre-N must have been performed in other places, like several lineages shared by Moroccans and Bushmen and such (that is not a smaller distance to the one from Ethiopia to Bangkok, mind you, actually it is more than double!)
There is no issue: people are not trees: we move.
"And almost without exception it can be assumed that those private lineages first appeared very near where they are now found".
That's a reasonable assumption under the principles of parsimony but in fact we have no idea and the rarer the lineage the less we can say of its history, we can just trace a straight line (or similar) between the region of the ancestor and the location where it's found. And that's what we do with the reconstructed origin of N and that of its ancestor L3. 6000 kilometers? Not such a huge distance, mind you.
"Their 'long tail' developed in situ"...
That's something you claim but that is not acceptable at all. The long or short "tail", "stem" more properly should be placed along the line between the ancestor and the coalescence location or actual documentation site. Of course uncertainty is huge and the intermediate steps may actually have happened anywhere capable of supporting human life but just spreading them evenly along the shortest possible migratory line fulfills the demands of goddess Parsimony and her mighty weapon, Occam's Razor.
Any other option, especially those that go away from this simplest possible reconstruction, demand evidence, at least indications...
"[N] is no more closely related to M than it is to the other L3 haplogroups as far as I'm aware. It's expansion is therefore almost certainly independent of M's".
There's no reason to believe such thing. It might be or not but the dotted line between Addis Abbaba and Mumbai (pre-M) and the one to Bangkok (pre-N) overlap, notably if we force them out of the Ocean (as must have been the case). Do you need me to send you a drawing or can you do that yourself?
"Unless you're yet again postulating genocide".
I don't even think that they were two different populations, just that while some "MN" groups (incidentally all M) expanded through South Asia, other "MN" subgroups, maybe those with greater boater or pioneer inclinations or some delay in development or whatever, went towards East Asia, where we find them side by side. What is a bit odd is that of all those Eastern "MN" lineages only N shows a marked star-like structure but that's also true in South Asia for M4"67 and nobody makes a fuzz about them.
Whatever happened anyhow, we cannot reconstruct the details, only narrate the highlights and that's not worth this endless discussion.
Back to one of your comments from yesterday:
ReplyDelete"Not true, Terry: the lineages are all South Asian (and not East Asian) in all cases and by no means restricted to minority populations like the Munda".
Yes, those lineages are South Asian, just as many other lineages are SE Asian. In neither case does that provide evidence as to which region the lineage as a whole coalesced in. In fact the South Asian lineages have not at any stage moved into SE Asia but a couple of SE Asian lineages have moved into Northeast India. F is present in Arunachal Pradesh, for example see:
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC2588665/
"The presence of haplogroups B5a and F1 in the Tharu of Eastern Uttar Pradesh (near the Indo-Nepal border) points to past human movements from East Asia to India, perhaps through Nepal".
And wiki claimes that R11 is present in Rajasthan:
http://en.wikipedia.org/wiki/Haplogroup_R_%28mtDNA%29
So we see a suggestion that it is easier to enter India from the east than it is to move from India to the east.
"I could do that, in fact I have done it in the past but you have not taken due note"
ReplyDeleteCould you please direct me again to where you did that. I'm sure your regions will not differ from the ones I have used.
"we see many examples every day of very small private lineages who are alive and well, often have been for tens of thousands of years".
Agreed. But can you show that they have traveled thousands of miles from where they coalesced? I'll bet not.
"We also see long migrations like pre-N must have been performed in other places, like several lineages shared by Moroccans and Bushmen and such"
Examples? I agree it's possible for two branches of a particular haplogroup to be widely separated from each other. But that is usually a product of unsuitable habitat between the two regions, or later expansion of other haplogroups that have separated a previously widely distributed haplogroup. We have no examples of a haplogroup dramatically appearing miles from its supposed origin, least of all a haplogroup with a long tail.
"in fact we have no idea and the rarer the lineage the less we can say of its history, we can just trace a straight line (or similar) between the region of the ancestor and the location where it's found".
I agree to some extent. But the straight lines you trace are between where a haplogroup is now found and where you would prefer it to have coalesced, not from where we 'know' it coalesced.
"That's something you claim but that is not acceptable at all".
It is surely extremely unlikely that a haplogroup could develop a long stem while on the move through habitat where it would be expected to remain for some time, establish a population branch and so diversify.
"The long or short 'tail', 'stem' more properly should be placed along the line between the ancestor and the coalescence location or actual documentation site".
But again you consistently 'make up' a coalescence location for no other reason than it fits your belief. I feel certain that if the evidence can be explained without making things up it is far more likely to be correct than if you have to postulate scenarios that may or may not have happened.
"Any other option, especially those that go away from this simplest possible reconstruction, demand evidence, at least indications..."
Exactly. And here you go making up all sorts of scenarios without any evidence at all.
"It might be or not but the dotted line between Addis Abbaba and Mumbai (pre-M) and the one to Bangkok (pre-N) overlap"
You're making unjustified assumptions again. Why do you insist that N coalesced in Bengal?
" What is a bit odd is that of all those Eastern "MN" lineages only N shows a marked star-like structure"
That is so 'odd' that it hardly bears considering. Why would N not have participated, at least to some extent, with M's colonisation of South Asia?
"but that's also true in South Asia for M4"67 and nobody makes a fuzz about them".
Nobody makes a fuss because it's perfectly obviousl what happened. Its parent haplogroup, M, was certainly present in India before M4''67's expansion.
"maybe those with greater boater or pioneer inclinations or some delay in development or whatever, went towards East Asia, where we find them side by side".
Maybe, maybe, maybe. But maybe not.
We have been through this matter of 'stems' before but here goes:
ReplyDelete"but in fact we have no idea and the rarer the lineage the less we can say of its history, we can just trace a straight line (or similar) between the region of the ancestor and the location where it's found".
I've checked through the basal mt-DNAs with stems as long or longer than N's. Here's what I found:
The haplogroup with the longest stem is R1 with 15 mutations. Whether it coalesced in South or West Asia it is reasonable to suppose that over the period of those 15 mutations it did not travel any huge distance from where it coalesced from basal R.
Next we find R23 with 14 mutations and R14 with 13 mutations. R23 straddles Wallace's Line and R14 is present in the Lesser Sunda Islands, as well as in New Guinea in the east and the Nicobar Islands in the west. You know very well I accept that these haplogroups have not moved very far at all from where they coalesced from basal R over the period of those 13/14 mutations. We also have Jordanian (?) R3 with 13 mutations from basal R. The presence of other basal R haplogroups nearby shows that basal R reached nearby regions so it is reasonable to suppose that R3 did not move far over the period of those 13 mutations.
We then find N8 in South China also with 13 mutations. We both agree that basal N was certainly present in that region whether it originated there or not, and so we should presumably agree that N8 did not move far at all for the period of those 13 mutations.
Then Sumatran M26 with 11 mutations. Basal M haplogroups are spread from India to Australia and so again it is reasonable to assume M26 did not move once it coalesced from basal M.
Then Australian N14 with 10 mutations. The presence of other basal N haplogroups in Australia hints strongly that N14 coalesced there. Again it did not move over the period of those 10 mutations.
Next we have M77 in Thailand with 9 mutations and M10, possibly from Vietnam, with 8 mutations. These two haplogroups almost certainly coalesced from basal M in the region they are found today and so didn't move over the period of those mutations.
We then find Northeast Indian R7 with 8 mutations. Whether R coalesced in South or Southeast Asia I'm sure we would agree that R7 probably coalesced from basal R in Northeast India. It didn't move over the period of those 8 mutations.
Then we come to Australian M15 with 7 mutations. Other basal M haplogroups are found in both Australia and new Guinea and so once more we can assume that M15 did not move after coalescing from basal M.
Now we're down to another Australian haplogroup, N13, with 6 mutations. Moved extensively over the period of those mutations? I doubt it.
Then we have a further 6 haplogroups with tails of 5 mutations. R22 in the Lesser Sunda Islands, R8 in South Asia, N21 in SE Asia, M76 in South China, M28 in Melanesia and A in East Central Asia. None of these haplogroups can convincingly be shown to have moved at all once they had coalesced from their respective basal haplogroups, although I know you insist that A has. Why should A be the only haplogroup to have moved from where it coalesced from its basal haplogroup? And let's not forget that N itself is 5 mutations from basal L3.
"We have been through this matter of 'stems' before"...
DeleteExactly. You imagine them one way and I do another (well, actually I imagine them no way because no evidence exists).
All you have to say are things like: ... "it is reasonable to suppose"... and stuff like that. Ironically you say the same for R-derivates from Jordan and Papua, and at least one of them (or all them probably) MUST have migrated from wherever R coalesced (IMO South Asia but would it be SE Asia it'd be the same).
The only "solution" that could fit your ideas is a "quantum uncertainty" model in which the R node, the R grandmother, was not here nor there but everywhere at the same time. And that is unacceptable.
Instead what I do is to narrow it by geographical (or geometrical) approximation, using (1) all nodes but also, for contrast and greater certainty, (2) only the short stemmed nodes (which coalesced first) and (3) even considering corrections towards the location of the ancestral node (L3 for N, N for R, etc.) These methods provide some rather precise points (1, 2 and 3, which may or not be near each other) and between the various results must almost necessarily be the real location of th actual R, N, M, etc. grandmas.
Long stems are just stretches of uncertainty between the rather precise estimate of geographical coalescence of the ancestor (R in your example) and the geographical location of its descendants (R subclades in West Asia and Oceania in your example). If these derived clades are near the ancestral origin, then it's probable that they coalesced near the "mother" (what is not necessarily good for success because the area was already populated), if they exist far away, then they coalesced far away and the stem... wherever: roughly in the dotted line between ancestral origin and actual destination - but we will never know for sure.
"You imagine them one way and I do another (well, actually I imagine them no way because no evidence exists)".
ReplyDeleteEvidence does exist actually. That evidence is the fact of the long tails. Such a tail can only develop as a result of drift knocking out other descent lines. That drift can basically only happen in a population of constant size, not in one that is expanding. Constant size suggests single region, not migrating population.
"Ironically you say the same for R-derivates from Jordan and Papua, and at least one of them (or all them probably) MUST have migrated from wherever R coalesced"
No. You seem to misunderstand basic population genetics. Those 'R-derivates' would not have moved all the way as separate haplogroups from wherever 'wherever R coalesced'. They would have arrived in the region where they are now found as undifferentiated R and then coalesced themselves in those regions. You are still stuck with this Garden of Eden belief.
"The only 'solution' that could fit your ideas is a 'quantum uncertainty' model in which the R node, the R grandmother, was not here nor there but everywhere at the same time. And that is unacceptable".
Ridiculous. It was onviously the R grandmother's clan that did the moving, not the daughters or grandaughters. I would have thought that was so simple to see, but obviously not if you are blindly committed to some other belief.
"(1) all nodes but also, for contrast and greater certainty, (2) only the short stemmed nodes (which coalesced first)"
That is looking at the evidence very selectively. You cannot ignore particular haplogroups just because they don't fit what you want to find. How do you decide on what constitutes the 'short stemmed nodes' and the others? What is your cutoff point? Surely all subclades began their differentiation from the parent haplogroup at roughly the same time, give or take a few thousand years. Haplogroups with long stems cannot be arbitrarily dismissed from your calculations, just because some of them don't fit what you're hoping to find.
"Long stems are just stretches of uncertainty between the rather precise estimate of geographical coalescence of the ancestor"
They are no more uncertain than are stems of any length. Under your system even haplogroups with a tail of just one mutation could have moved huge distances from where the parent haplogroup lived.
"Evidence does exist actually. That evidence is the fact of the long tails. Such a tail can only develop as a result of drift knocking out other descent lines. That drift can basically only happen in a population of constant size, not in one that is expanding. Constant size suggests single region, not migrating population".
DeleteActually some expansiveness is almost needed for small clades to survive (always Chaos allowing): in constant size small clades tend to vanish by mere drift and fixation on the majority clade tends to arise easily. Constant size is almost as bad for small clades as contraction.
Whatever the case, we do not know how many other small clades which did not survive existed in the past. Probably many, what we see now is just a remnant of that ancient diversity, gradually subsumed into nothingness by their more common relatives.
There is no determinant however for how long a small clade can survive or which history it may have gone through: it's just a huge lot of heads and not a single tail, at least not in that particular line. And that is simply not predictable: a long unbranched stem is a peculiar product of Chaos and not much can be said about Chaos that makes sense.
"They would have arrived in the region where they are now found as undifferentiated R and then coalesced themselves in those regions".
Not necessarily: they could also have accumulated mutations before arrival. IF the journey took some time and the founder group was small it's almost certain that they did. Only an instant teleportation, so to say, a very fast migration would allow for no mutations to have accumulated before arrival.
But, even in that case, arrival to destiny, normally in those times a favorable, maybe virgin, niche, would cause expansion. So I contend that, at least in most cases, these lineages only expanded (coalesced, formed the branching node that carries their name) upon arrival to destination, which was almost invariably a good place to live and prosper.
This last should apply to all clades that show sings of expansion (branching nodes); on the other hand, lineages with no obvious signs of expansion must have been all the time in "the Chaos zone", bordering extinction and never expanding and very little can be said about them, except being listed in the diversity counts.
So it has no relation whatsoever to how you imagine it: lines that did not branch actually provide almost no information (all the time squeezed probably, just lucky enough to survive) and lines that did branch provide info only upon branching (arrival to a good place to live).
"... You cannot ignore particular haplogroups"...
DeleteI did not say that. I said that in a second test I do focus on the older lineages but in the first test I count them all. I'm ignoring nothing, just looking at various layers of the information we have. Anyhow they tend to produce the same or very similar results in my experience, what is reassuring.
"If these derived clades are near the ancestral origin, then it's probable that they coalesced near the 'mother' (what is not necessarily good for success because the area was already populated) if they exist far away, then they coalesced far away and the stem... wherever: roughly in the dotted line between ancestral origin and actual destination - but we will never know for sure".
ReplyDeleteSurely once it's pointed out you will be able to see where your claim falls to the ground.
You're proposing that it took mt-DNA N14 mutations to do acheive what mt-DNA S did in one: move from Bengal to Australia. O was a little slower than S. That haplogroup took two muations to travel the distance, while the sluggard N13 was able to linger along the way for six mutations. That necessitates four migrations into Australia, just by mt-DNA N. And mt-DNA P too moved from South Asia to Australia in a single bound, just one mutation from basal R. Many people are reluctant to accept any more than one Paleolithic Wallace's Line crossing. Surely it is extremely likely that all five haplogroups arrived in Australia within a short time of each other.
In the other direction we have R0 and R2'JT emerging from South Asia over a period of just one mutation. Then U came along two mutations later followed twelve mutations later by R3, and later still by R1. That's at least three migrations west from South Asia for R alone. N suffers from a similar problem. It took N1'5 a period of just one mutation to emerge from India but assorted other N haplogroups took longer. X and N2 four mutations, while poor old N3 had to wait until it had accumulated 12 mutations (not to mention a number of control region mutations) en route from India.
Surely it is much more likely that all these different haplogroup expansions basically represent as few as four movements in all. Over extended periods perhaps, but certainly not a whole series of individual haplogroup migrations from a single region of origin: a Garden of Eden. Of course many individual haplogroups have undergone subsequent individual expansions of their own but others have remained in a single region and accumulated a long stem.
And let's not forget that the common ancestor of the three haplogroups R11'B6, B4'5 and R24 had managed to escape from India so rapidly that it was able to accumulate just a single control region mutation along the way.
"Surely once it's pointed out you will be able to see where your claim falls to the ground".
DeleteHow can a person who is so often so wrong can be so cocky?!
"You're proposing that it took mt-DNA N14 mutations to do acheive what mt-DNA S did in one: move from Bengal to Australia".
No. But the fact that N14 (which is so far just a "proposed", unconfirmed lineage anyhow) shows no signs of expansion suggest that it did arrive well after S and O and found no place to expand, remaining "private". Alternatively it was eaten by a grue, i.e. was very localized in a region that suffered a catastrophe like sudden drought or whatever. But most likely the first explanation: it never found a place to expand and is sort of a ghost from the past which survived only by luck.
"Many people are reluctant to accept any more than one Paleolithic Wallace's Line crossing".
It doesn't seem like anything I should consider: "many people" (do I look like I care about what "many people" may think?) "are reluctant to accept" (those "many people" look to me like a bunch of spineless conservatives with an overgrown amygdala anchored to some fetish that gives them a false sense of security) "any more than one Paleolithic Wallace's Line crossing" (any particular reason why? - just a rhetorical question of course, no need to answer: no true reason, just their panic to accept daring and genius in "primitive" people, is the real reason).
What prevented newcomers from forming new populations across semi-permeable barriers like Wallace Line, etc. was not their inability to cross them (they were able in all cases) but the fact that only few crossed at any given time and there were much larger populations at destination, which in the best case absorbed them, drifting out their lineages in many cases.
So N14 might have arrived at any time but in the early waves. Otherwise it would have thrived (and did not).
I'll leave it at that.
"How can a person who is so often so wrong can be so cocky?!"
ReplyDeleteBecause I am certainly correct on this matter at least, and you are wrong.
"what we see now is just a remnant of that ancient diversity, gradually subsumed into nothingness by their more common relatives".
I agree 100%, but that is irrelevant when we're just considering surviving haplogroups.
"So I contend that, at least in most cases, these lineages only expanded (coalesced, formed the branching node that carries their name) upon arrival to destination, which was almost invariably a good place to live and prosper".
On the contrary, if they had not originally arrived in 'a good place to live and prosper' they would not have survived in the first place. They would have arrived in the region as a result of the expansion of their parent haplogroup and then accumulated the tail over time. In many cases haplogroups have been able to expand once more when times are amenable, but this is after they've accumulated their tails.
"This last should apply to all clades that show sings of expansion (branching nodes)"
Yes, such as M, N, R. These haplogroups left separate clades to develop regionally once the parents had expanded expanded widely. The same has happened with other haplogroups derived from these basal ones, but expansion is associated with diversification not the development of a long tail.
"lineages with no obvious signs of expansion must have been all the time in 'the Chaos zone', bordering extinction and never expanding"
At last, 'never expanding'.
"very little can be said about them, except being listed in the diversity counts".
On the contrary. Because they have never expanded they can tell us rather a lot.
"But the fact that N14 (which is so far just a 'proposed', unconfirmed lineage anyhow) shows no signs of expansion suggest that it did arrive well after S and O and found no place to expand, remaining 'private'".
There could be any number of reasons why it remained a private lineage.
"Not necessarily: they could also have accumulated mutations before arrival".
So extremely unlikely that it is not worth considering. I think I've tracked down one of the L haplogroups you mentioned a few days ago: L1c3b2, with twelve mutations from L1c3b'c. Its relations are concentrated in West Africa, specifically Gabon according to your excellent blog on African mt-DNA haplogroups. It's having reached southern Africa is easily seen as part of the Bantu expansion. Its presence in Morocco is possibly a result of contact via Saharan trading routes. Certainly its modern distribution is a product of movement AFTER the haplogroup had accumulated its long tail. The haplogroup cannot be shown to have moved a centimetre while it accumulated that tail.
So what about more recently coalesced haplogroups? I have accumulated information for haplogroups involved in Southeast and East Asian populations. B5b, 8 mutations from B5, is found in Laos/Vietnam. Almost certainly B5b coalesced in that region. B4f, 7 mutations from B4, is found in South China somewhere. Almost certainly very near where it began accumulating its tail. R11, 7 mutations from R11'B6, is found in South China (especially amoung the Lahu of Yunnan). R11 is simply the western version of R11'B6 just as R24 is the eastern, or Philippine, version of R11'24'B'. Almost certainly R11 did not move a centimetre once it began the proccess of accumulting its long tail.
And so on. You have not yet provided a single example of a haplogroup that may have developed any sort of tail while on the move.
A place being "a good place to prosper" is mot a mere function of its raw ecological or economical excellence but of how many people it has to sustain (relative to the extant technological capacities, which is mostly a variable we can ignore). So let's say that South Asia could support 20,000 people comfortably (c. 4x what Bocquet-Appel estimated for Europe before the "Magdalenian revolution", avg., it could well be 100,000 but your take) the 20,001 is squeezed in and so is maybe the 20,500 but soon tensions arise and people have to either kill each other or migrate (or both).
DeleteIt's not something strict or absolute (people adapt and find maybe new sources of food, climate or human pressure may take a toll on those resources, etc.) but very generally a tribe that can will protect their territory from outsiders either by means diplomatic or bellicose. As resources become more scarce, bellicosity becomes more important, logically.
So your claim:
"On the contrary, if they had not originally arrived in 'a good place to live and prosper' they would not have survived in the first place".
... seems to lack substance because the emigration probably happened in a later stage when resource began to be scarce for reason of relative overpopulation (or whichever).
To put a modern example, my parents may have lived in a golden age of jobs when everyone had not one but even often two jobs and social security promises but today 25% are unemployed and social security is being demolished. Or you could also say that Europe is a nice place to live. It is... but millions of Europeans emigrated in hope of better lives in the not so distant past because for them particularly it had become a rather oppressive and hopeless environment. Etcetera.
"On the contrary. Because they have never expanded they can tell us rather a lot".
Not at all: all in them is a random fluke and flukes are not statistically significant. Only pondering them in group we may reach a zone of some statistical significance but in isolation they tell us nothing but their present day unstable status.
"So extremely unlikely that it is not worth considering".
You have no reason to make such claim. Non-branching stems may have accumulated their line of mutations anywhere, absolutely anywhere (even if for parsimony reasons we generally assume that in the journey between ancestor's origin and end of travel at present time). It's only when there are branching nodes when we get some information in form of imaginary "arrows" pointing to the respective destinations of the subclades in a hierarchical manner.
...
...
Delete"I think I've tracked down one of the L haplogroups you mentioned a few days ago: L1c3b2, with twelve mutations from L1c3b'c. Its relations are concentrated in West Africa, specifically Gabon... Its presence in Morocco is possibly a result of contact via Saharan trading routes."
Actually I was thinking of L0a1b1. But you may be right on this one because they are both not described as Khoisan but "South Africa" (so they could be Bantu). Still a bit mysterious.
"Certainly its modern distribution is a product of movement AFTER the haplogroup had accumulated its long tail".
I doubt I used this lineage as example re. your long tail misconceptions because even if we don't know of its phylogenetic branching (there may be one) the geographic branching is such that it is indeed suggested. Also my actual lineage L0a1b1 has a very short tail (only one known mutation in the stem from L0a1b).
I don't understand why you bring that here. You force me to address the most obscure elements of your confusion in what I deem a total waste of time.
"Almost certainly R11 did not move a centimetre once it began the proccess of accumulting its long tail".
That's because of parsimony (origin = destination) not because of the stem (not "tail": you are not born out of your tail, if you have one, but you are from a stem or umbilical cord).
"You have not yet provided a single example of a haplogroup that may have developed any sort of tail while on the move".
All them but of course cannot be demonstrated. What I say is that they accumulate the mutations (for reasons of parsimony only) between the ancestor's origin present day destination. If they are coincident, as in your example, the carriers did not move much probably; if they are not, as in the many R-derived examples mentioned previously or as in N and M in relation to L3, then the carriers did move (obvious) and may have accumulated the mutations at any point between origin and destination. We cannot know for sure: there is no way we can know that.
"Actually I was thinking of L0a1b1. But you may be right on this one"
ReplyDeleteOnly that one? I defy you to point out one where I'm wrong.
"I don't understand why you bring that here. You force me to address the most obscure elements of your confusion in what I deem a total waste of time".
I bring the matter up because once you can see what I'm getting at you will be better able to interpret the distribution of haplogroups in Europe, which I think is your main interest. Interpret the evidence of a long tail FIRST as indicating a long period in the same region. Examine alternative interpretations only once that is absolutely impossible. I am yet to find an example of this last.
"What I say is that they accumulate the mutations (for reasons of parsimony only) between the ancestor's origin present day destination".
And what I say is that they accumulate the stem once their 'parent' haplogroup has reached the region where they are now found. In other words basal mt-DNA R had reached the region where R11 is now found before the first mutation on the stem had happened. Of course at that expansion R was already a little diverse but that diversity increased dramatically as the haplogoup expanded in numbers and distribution.
"if they are not, as in the many R-derived examples mentioned previously or as in N and M in relation to L3, then the carriers did move (obvious) and may have accumulated the mutations at any point between origin and destination. We cannot know for sure: there is no way we can know that".
It is possible that we cannot be absolutely sure, but we can be fairly certain that the 'stems' developed in a small, isolated population unlikely to be moving very far at all.
"A place being 'a good place to prosper' is mot a mere function of its raw ecological or economical excellence but of how many people it has to sustain (relative to the extant technological capacities, which is mostly a variable we can ignore)".
The first arrivals are very unlikely to immediately fill the whole ecosystem. The population would usually establish itself then members will peel off and move. These ones that move will often develop into a subclade of the 'original' haplogroup but if they are able to continuously expand we will find a star-like structure. If unable to expand further they will start developing a long stem.
"And what I say is that they accumulate the stem once their 'parent' haplogroup has reached the region where they are now found".
DeleteAnd I say that that claim is totally absurd and that you are fundamentally wrong in this.
And also that I'm bored of arguing in circles. I have explained all and more than I could: if you can't understand it is not my problem anymore.
"As resources become more scarce, bellicosity becomes more important, logically".
ReplyDeleteAnd that situation is well accepted by Pacific Island archeologists. The first arrivals harvested the most easily accessible resources to start with, expanded in numbers and then, when the resources tended to run out, indulged in warfare. Fortifications do not appear on each island until some time after original occupation.
"So your claim ... seems to lack substance because the emigration probably happened in a later stage when resource began to be scarce for reason of relative overpopulation (or whichever)".
At which point the haplogroup would have ceased accumulating a long tail and begun diversifying again.
"but millions of Europeans emigrated in hope of better lives in the not so distant past"
That option was not usually open to Paleolithic people. They would usually have been attacked by members of other tribes, so would not have survived to produce offspring in the new region. Even if accepted into another tribe their haplogroup would be such a minority that it would be unlikley to survive many generations. Unless the incoming individual had some huge advantage over the locals. Suiperior technology, for example, but even that would probably be passed to other members of the new tribe.
"Non-branching stems may have accumulated their line of mutations anywhere, absolutely anywhere"
I keep trying to get through to you that it is extremely unlikely that they would accumulate those stems while on the move though.
"It's only when there are branching nodes when we get some information in form of imaginary 'arrows' pointing to the respective destinations of the subclades in a hierarchical manner".
The 'branching nodes' give us some idea of where the particular expansion began but the only difference between a branchinmg node and a long stem is that the first expanded independently of its parent and the second did not expand. The 'points' of each tell us much the same thing.
"At which point the haplogroup would have ceased accumulating a long tail and begun diversifying again".
DeleteNo! The mother lineages' numerical dominance would not allow them to thrive past a point (in the expansion or towards the stabilization). Expansion, notably the sudden and fast expansion of the star-like type, only happens when a new niche opens (either because of colonization of new areas or other reasons):
1. Arrival to an open niche and immediate expansion (founder effects and formation of several/many derived lineages from these).
2. Stabilization and fixation (natural pruning or "drift-out" of less-lucky minor lineages).
That's the process.
With luck some of the smaller lineages may survive the pruning in "private" form to make a new founder effect elsewhere (usually the emigrants are the underdogs at the motherland) along with descendants of the more dominant clades. That's why some lineages appear to "vanish" (or rather just be relatively rare) in some areas only to reappear elsewhere with just less obvious or even invisible links in between.
"I keep trying to get through to you that it is extremely unlikely that they would accumulate those stems while on the move though".
I keep telling you that I disagree.
First of all because "on-the-move" is a quite relative term for semi-nomadic peoples as were all hunter-gatherers. Also there's no objective reason rejecting possible intermediate stops: "on the move" maybe but they had to sleep, eat and collect resources, what probably meant long stays, probably trans-generational ones, in intermediate areas that did not allow them to expand wildly but did keep them alive. In all those many stops through the millennia they must have accumulated mutations before arrival.
A very rough estimate is one coding-region mutation per each several (1-3?) millennia. That means that if M accumulated three mutations before finding a niche to expand, it was maybe 3-10 millennia in Arabia (not much considering that pre-Jurreru phase in Arabia/Palestine extends for maybe as much as 50 Ka).
What does not make sense at all is that L3 would migrate right away from the East Africa to India as such undefined L3 root and then, instead of expanding wildly right away, it waited for 3-10 thousand years accumulating mutations before expanding at all, only to make Terry happy.
No way!
If you can't explain M, you can't explain anything. And your weird conjecture cannot explain M.
"No! The mother lineages' numerical dominance would not allow them to thrive past a point (in the expansion or towards the stabilization). Expansion, notably the sudden and fast expansion of the star-like type, only happens when a new niche opens (either because of colonization of new areas or other reasons)"
ReplyDeleteExpansion leads to diversification. It follows, therefore that lack of expansion leads to lack of diversification. In a small population mother lineages would often tend to be replaced by daughter lineages. But new daughter lineages also form as the haplogroup diversifies during expansion of course. And tails cease to accumulate.
"With luck some of the smaller lineages may survive the pruning in 'private' form to make a new founder effect elsewhere"
But in most cases those 'smaller lineages' will have formed from a parent lineage once the parent lineage had reached that region.
"1. Arrival to an open niche and immediate expansion (founder effects and formation of several/many derived lineages from these)".
We see the phenomenon clearly in mt-DNA B, the haplogroup I am perhaps most familiar with. And we are fairly sure of both the sequence and the timing of its expansion. B4a is most likely a mainland East/Southeast Aasia haplogroup although B4a(xB4a1) reaches Polynesia. B4a1a is present on both Taiwan and the Philippines, but the derived B4a1a1 is not present in Taiwan but is present in the Philippines. It must have coalesced there. B4a1a1a is known as the 'Polynesian motif', found from Madagascar to new Zealand. However madagascar has B4a1a1a2, not found elsewhere, while Polynesia has B4a1a1a3, also not found elsewhere. Both B4a1a1a1 and B4a1a1a4 have been found in both Melanesia and Polynesia. Although none of these subclades of B4a1a1 have a long tail it is reasonably easy to see that they did not coalesce in the Philippines but in the regions where they are now found.
You will no doubt be inclined to say, 'But these are oceanic haplogroups'. But even in the Eurasian landmass expansion would not have been like ink through blotting paper. People would have occupied the most desirable locations first. Whether these desirable regions were separated by swathes of difficult territory or just a few kilometres of slightly less desirable habitat the effect would have been much the same as a spread through islands.
"probably trans-generational ones, in intermediate areas that did not allow them to expand wildly but did keep them alive. In all those many stops through the millennia they must have accumulated mutations before arrival".
Any small group migrating through such regions of isolated habitats each of limited extent would be unlikely to survive long enough to leave any descendants at all. Inbreeding with the group would have devastating consequences within a few generations for a start.
"That means that if M accumulated three mutations before finding a niche to expand"
No. It accumulated three mutations before it was ble to expand.
"it was maybe 3-10 millennia in Arabia"
Very likely so, I'd say. Or more likely somewhere near the Persian Gulf, perhaps east of it.
"What does not make sense at all is that L3 would migrate right away from the East Africa to India as such undefined L3 root"
I have never suggested any such thing. It is just your imagination running riot again. However undifferentiated M almost certainly entered India and moved as far as Zomia. After it had expanded across this whole region the regional variations in haplogroup M were able to form and came to dominate the parent haplogroup: M.
"If you can't explain M, you can't explain anything. And your weird conjecture cannot explain M".
I think I just did expalin haplogroup M, and what 'weird conjecture' are you accusing me of?
"Although none of these subclades of B4a1a1 have a long tail it is reasonably easy to see that they did not coalesce in the Philippines but in the regions where they are now found".
DeleteThe rhythm you attribute to those mutations is too fast. They probably existed once in Philippines (or wherever) and were later drifted out (or still exist infrequently enough not to be detected by sampling).
If, as it is the case with your example, there was a founder effect, the almost certain case was that the haplogroup arrived already derived in that founder effect form (B4a1a1a in Polynesia, for example). Otherwise how can you explain the prunning of all the other B4a1a1* that must have existed right after the colonization? Never mind the excessively fast mutation rate...
I said: "... M accumulated three mutations before finding a niche to expand"
You replied: "No. It accumulated three mutations before it was able to expand".
Which is exactly what I was saying (take out the "no") because it was unable to expand ONLY before reaching destination. Once it reached its expansion zone (surely South Asia) it did expand because it was suddenly able to do it (only then and not before nor, so strongly at least, later either).
"However undifferentiated M almost certainly entered India and moved as far as Zomia".
Or not. Probably the many clades that created founder effects East of Bengal arrived there already formed (but in small numbers, hence the founder effect).
You'd ask: then why don't we find them in India? Because they were tiny back then and were pruned by the expansion of other more numerous, dynamic or lucky "sister" lineages. That's how drift works.
You'd then say: always appealing to founder effect and drift. And I'd say: because that's how things actually work, spare me.
"I think I just did expalin haplogroup M"
You "expalined" nothing.
"The rhythm you attribute to those mutations is too fast. They probably existed once in Philippines (or wherever) and were later drifted out (or still exist infrequently enough not to be detected by sampling)".
ReplyDeleteOn what evidence do yopu base that claim? I'm sure you just believe such to be so because it suits what you want to believe.
"Otherwise how can you explain the prunning of all the other B4a1a1* that must have existed right after the colonization?"
Exactly the same way as I explain other cases where a daughter haplogroup has replaced a parent haplogroup in a small population. I accept the fact that you don't accept that fact, so I should abandon all hope.
"because it was unable to expand ONLY before reaching destination".
My guess, and it is only a guess, is that a change in climate allowed the expansion from a previously limited region.
"Once it reached its expansion zone (surely South Asia) it did expand because it was suddenly able to do it"
Quite probably its expansion in India was related to the same change in climate that allowed M to reach India in the first place.
"Probably the many clades that created founder effects East of Bengal arrived there already formed"
Some, possibly. But most NE Indian clades are found only in NE India or further east. However we do have one clade that stretches right across India from Arabia to Laos. That haplogroup is M1'20'51. M1 has a tail of four mutations, possibly because it represents the branch of M that continued living in the small region M as a whole had been confined to. M51 has a tail of six mutations but surely it is reasonable to suppose that it developed that tail somewhere between Laos and Nepal, before exanding to the other region. And M20 has a massive tail of fourteen mutations, almost certainly developed once it had reached the South China/Vietnam region.
"then why don't we find them in India? Because they were tiny back then and were pruned by the expansion of other more numerous, dynamic or lucky 'sister' lineages".
In which case we would surely expect to find some of these 'numerous, dynamic or lucky sister lineages in NE INdia. We don't. What we find with M is basically two geographic regions of expansion. A Central Indian one and a NE Indian one. There is little overlap except in the eastern coastal regions of India which is easily explained as being the product of later movement from both regions. That distribtion strongly suggests that mt-DNA M did not arrive in India via the coast.
A few comments above you agreed with me that mtDNA coding region mutations only seem to accumulate at the slow rate of 1000-3000 years each. It's a very rough approximation of course but I understand that the Oceanic expansion was quite faster, at least faster between Philippines and wherever the "Polynesian motif" founder effect took place: a matter of centuries or even decades. So there's an apparent contradiction here.
Delete"Exactly the same way as I explain other cases where a daughter haplogroup has replaced a parent haplogroup in a small population".
It does not make sense: the population was expanding fast and into many different smaller niches (islands, archipelagos): different sublineages should have survived in each destination if you'd be right (which you aren't).
"My guess, and it is only a guess, is that a change in climate allowed the expansion from a previously limited region".
DeleteI'm rather inclined to think that the change of climate pushed migration into unknown lands. After all we are before a colder and drier period at the end of the Abbasia Pluvial (if the migration effectively happened c. 80 Ka., as suggested by Petraglia's data), a period of contraction and not expansion for mere climatic reasons. It was the pump process which triggered the migration and therefore, only indirectly, the expansion... by making the carriers of M to arrive to South Asia, where they expanded happily. This geography of the expansion is demonstrated by the very geographical distribution and star-like structure of the haplogroup itself.
"In which case we would surely expect to find some of these 'numerous, dynamic or lucky sister lineages in NE INdia".
We do.
"We don't".
We do, I insist.
"What we find with M is basically two geographic regions of expansion. A Central Indian one and a NE Indian one".
First news I have of such structure. AFAIK, most South Asian large M sublineages are pretty much widespread through the subcontinent.
"A few comments above you agreed with me that mtDNA coding region mutations only seem to accumulate at the slow rate of 1000-3000 years each".
ReplyDeleteI thought you had trouble with molecular-clockology. Only when it suits you obviously. I am agnostic on the subject. I see no reason why the mutation rate should be constant in the first place.
"I'm rather inclined to think that the change of climate pushed migration into unknown lands".
That is possible. But climate deterioration would tend to force people into regions that remained habitable. Over time these regions would become isolated and it would become impossible to leave them and move to more desirable locations. Extinction would follow.
"First news I have of such structure. AFAIK, most South Asian large M sublineages are pretty much widespread through the subcontinent".
Not so. Even your own list says otherwise:
http://ourorigins.wikia.com/wiki/MtDNA_haplogroup_M
And you might like to compare your list with this one:
http://ourorigins.wikia.com/wiki/Mt_M_west_to_east
No contradictions between the two as far as I can see.
I also thought you would come to your senses and have a constructive debate some day instead of just opposing everything word after word.
Delete...
"But climate deterioration would tend to force people into regions that remained habitable. Over time these regions would become isolated and it would become impossible to leave them and move to more desirable locations. Extinction would follow".
... if those regions also became inhabitable and all the rest you say would apply. But that's not what happened in South Asia. So why to speculate in a vacuum when you can look at the real thing?
"http://ourorigins.wikia.com/wiki/MtDNA_haplogroup_M"
Have you even looked at the gallery at the bottom? All six haplogroups represented there more or less cover homogeneously all or most of the subcontinent. What the heck are you talking about? And they can't be the only ones, of course.
"I also thought you would come to your senses and have a constructive debate some day instead of just opposing everything word after word".
ReplyDeleteNow you know exactly how I feel.
"All six haplogroups represented there more or less cover homogeneously all or most of the subcontinent".
No they don't. They are certainly not evenly distributed through South Asia. And none of them appear to have spread from Northeast India. Half of them haven't even reached there. In fact I used those maps to make my own entry at Wikimedia. M2 is concentrated at the mouth of the Ganges with a second centre further down the coast around the mouth of the Godavari, up which it appears to have spread as far as Gujarat. M3 is the most widespread but it is centred in western Madhya Pradesh, and its subsequent spread appears to have been halted by the Ganges. M25 is centred on the west coast, with some indication of a concentration on the Narmada River valley, and a presence in the Punjab. M6 is particularly interesting as it is spread along the east coast of India but also crosses the Ganges to spread around the Bay of Bengal. The remaining two are part of M4''67. M4a at the mouth and the headwaters of the Indus with an interesting outlier on the southeast coast at the mouth of the Godavari. M18 at the mouth of the Godavari with a strong presence in Rajasthan. It also has a minore presence at the Ganges Delta but that could easily be the product of a later migration. In fact M4a and M18 don't overlap in their distribution at all except for at the mouth of the Godavari. The haplogroups obviously did not originate in some Garden of Eden and all spread from there fully developed.
The Mutation for M25 is always directed to Kerala. And, it is in Punjab.
DeleteYou got it all wrong. Some of these castes that you classified as Artisan and Warrior class have so many farmers. And some of the Dry Land farmers have aristocrats and warriors as well. What you don't know is not all people in one caste belong to only a warrior caste or a farmer caste. Within the caste their are sub castes. Make sure you do your research properly. I have lived in the region for decades so I know the local history and folklore of that region.
ReplyDeleteI did nothing, Guna, the authors of the study did. Please address your grievances to them (link is at the beginning of the entry, in case you missed it).
DeleteI merely discussed their work.
When you look at ezhavas and mukuvars they were farmers as well, their occupation was coconut farming and fishing. The kallars and nadars have warriors. The nadars come from an assortment of other castes and classes which was later years catagorized as nadars. So the authors are mostly incorrect of these castes. When observing the ydna haplogroup its most likely the L dna haplogroup who brought their proto dravidian ways to South India. The H is found its presence all over India, R1a has its strong presence from west asia most likely their immigration to India took place due to the islamic conquest. The J dna has it presence in south india before the R1a. The R1b dna is most likely western european influence, due to the spice trade.
ReplyDeleteAnyone have info on the U8a and C1a contribution? For the Ancient Brahmi related Tamils in Egypt. As for our rare T2b6b 7391C variety, the paternal side shows a Canarian related contribution.
ReplyDeleteBe back in a short while..Thanks!